Figure 7 |. HMGB1 mediates crosstalk between FBP1-deficient hepatocytes and HSCs.
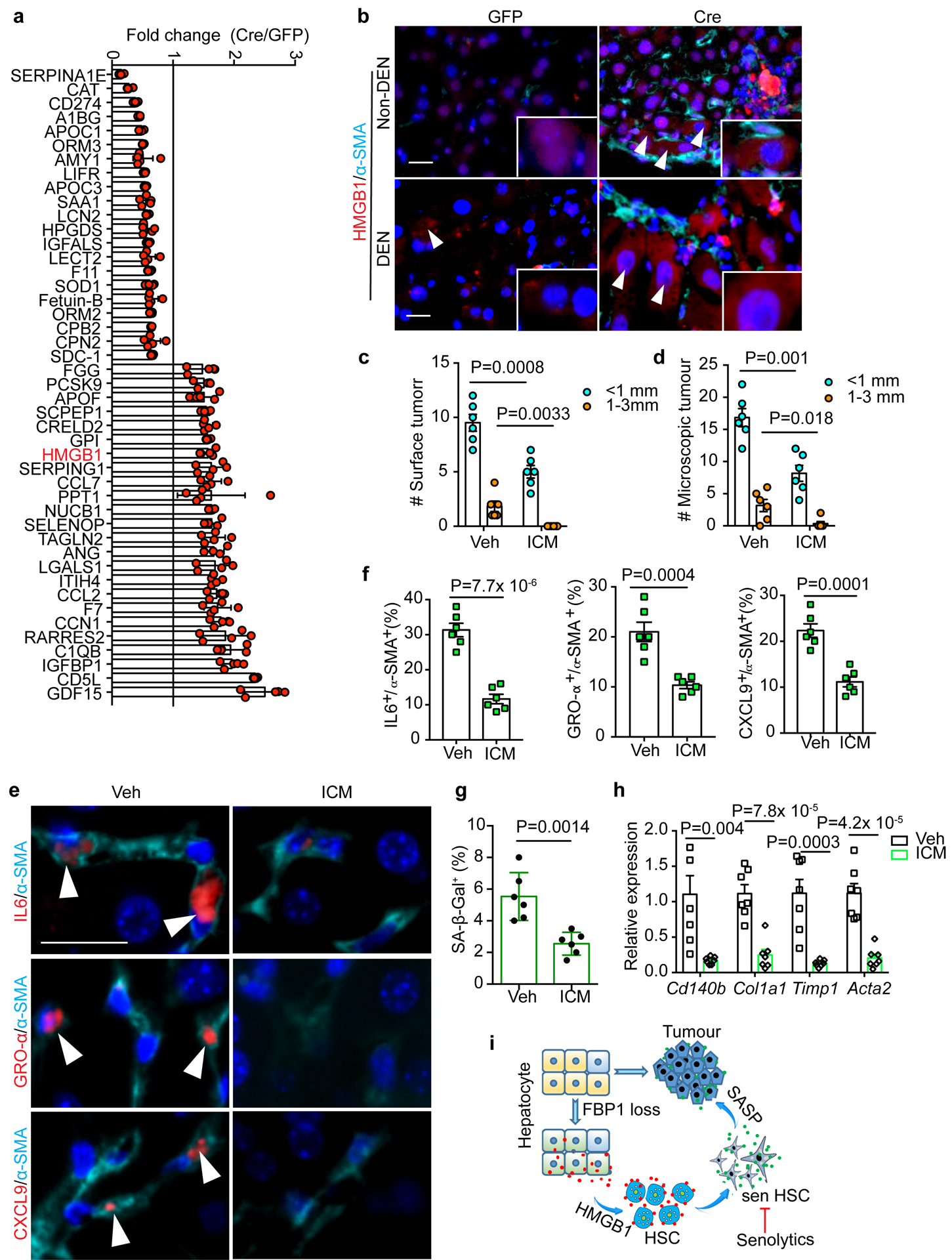
a, Fold change of 45 secretory protein abundances in Cre relative to GFP hepatic CM (adjusted p<0.05). 5 biological replicates from 3 mice of each cohort. b, Representative IF staining of HMGB1 and α-SMA in 24-week mouse liver sections (n=3 independent experiments). Scale bar: 50 μm. White arrowheads indicate cells with cytosolic HMGB1 staining. c, d, Quantification of surface tumour (c) and microscopic tumour (d) number and size distributions in Veh (n=6) and ICM (n=7) DEN/Cre mice. e, f, IF staining (e) of IL6+/α-SMA+, GRO-α+/α-SMA+ and CXCL9+/α-SMA+ cells and quantification (% of α-SMA+) (f) in mouse liver sections from Veh (n=6) and ICM (n=7) DEN/Cre cohorts. Scale bar: 50 μm. White arrowheads in (e) indicate cells with double positive staining. g, Quantification of SA-β-Gal staining (% of cells) in 24-week Veh (n=6) and ICM (n=6) DEN/Cre mouse liver sections. h, qRT-PCR analysis of fibrotic gene expression from 24-week Veh (n=6) and ICM DEN/Cre (n=5) mouse livers. i, Working model for liver tumour promotion by hepatic FBP1 loss. Hepatic FBP1 loss disrupts liver metabolism leading to ER stress and a distinctive hepatic secretome; secreted HMGB1 as one mediator activates HSCs; HSCs undergo senescence (sen HSC) and promote tumour progression through a SASP. Graphs in c, d, f, g and h show mean ± SEM, and P values were calculated using a two-tailed t-test. Numerical source data are provided in Statistic Source Data Fig. 7.
