Figure 3.
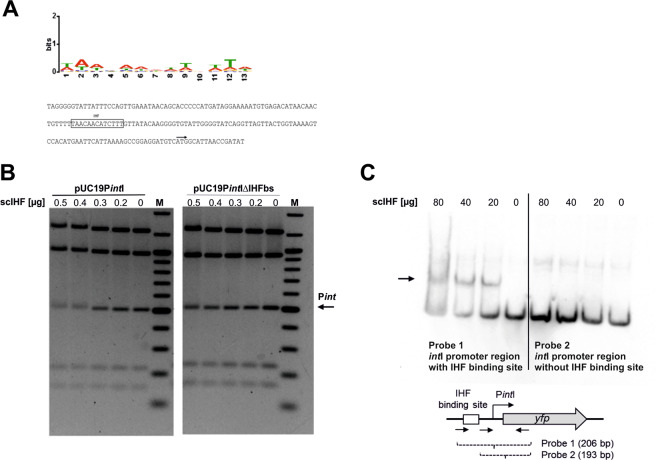
IHF binds to a high affinity binding site in the intI promoter region. (A) Sequence logo used to computationally predict IHF binding sites in the vicinity of intI-VI (up) and nucleotide sequence upstream of the intI gene (down). The binding site sequence logo was created using MEME39 and the experimentally identified IHF binding sites listed in RegulonDB38. Annotated are the main features of the intI gene: predicted IHF binding site (box), start codon of the coding sequence of intI (black arrow). (B) Analysis of scIHF binding to the intI upstream region by electrophoretic mobility shift assay (EMSA). Single-chain IHF bound specifically to the promoter fragment (PintI) only in the presence of the IHF binding site (left), but not when the binding site was absent (right). M, DNA size marker. For this figure, the original gel has been cropped and the arrangement of the two parts of the gel representing either pUC19PintI or pUC19PintI ΔIHFbs has been reversed. The original agarose gel is provided in Supplementary Figure S6. (C) Digoxygenin-labeled probes comprising the intI upstream region with or without IHF binding site (25 ng) were incubated with or without purified scIHF (80 µg, 40 µg, 20 µg; 0 µg). Single-chain IHF bound specifically to the promoter fragment (PintI) only in the presence of the IHF binding site (left), but not when the binding site was absent (right).