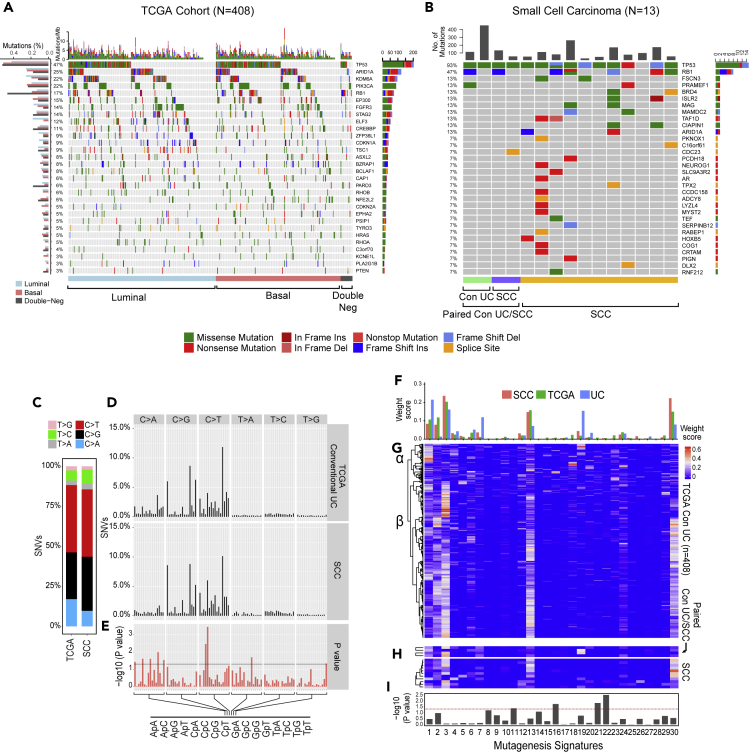
Figure 1.
Mutational Landscape of SCC
(A) Mutational landscapes among the molecular subtypes of 408 muscle-invasive bladder cancers from the TCGA cohort showing the frequency of mutations in individual tumors and somatic mutations for significantly mutated genes. The frequencies of mutations of individual genes in the luminal, basal, and double-negative subtypes are shown on the left. Bars on the right show the numbers of specific substitutions for individual genes.
(B) Mutational landscapes of 13 cases of SCC and 2 paired samples of precursor conventional UC showing the frequency of mutations in individual genes and somatic mutations for significantly mutated genes. The frequencies of mutations of individual genes are shown on the left. Bars on the right show the numbers of specific substitutions for individual genes.
(C) Composite bar graphs showing the distributions of all nucleotide substitutions in two sets of samples corresponding to the TCGA cohort and SCC.
(D) Proportion of single-nucleotide variants (SNVs) in specific nucleotide motifs for each category of substitution in two sets of samples as shown in (C).
(E) False discovery rate (FDR) for specific nucleotide motifs in two sets of samples as shown in (C).
(F) Average weight scores of mutagenesis patterns in three sets of samples corresponding to the TCGA cohort, paired precursors conventional UC, and SCC.
(G) Weight scores of mutagenesis patterns in individual tumor samples of the TCGA cohort.
(H) Weight scores of mutagenesis patterns in SCCs and paired precursors conventional UCs.
(I) Statistical significance of mutagenesis patterns (p value) in SCC compared with conventional UC. For (E) and (I), p value was calculated using Wilcoxon rank-sum and Kruskal-Wallis tests, respectively.