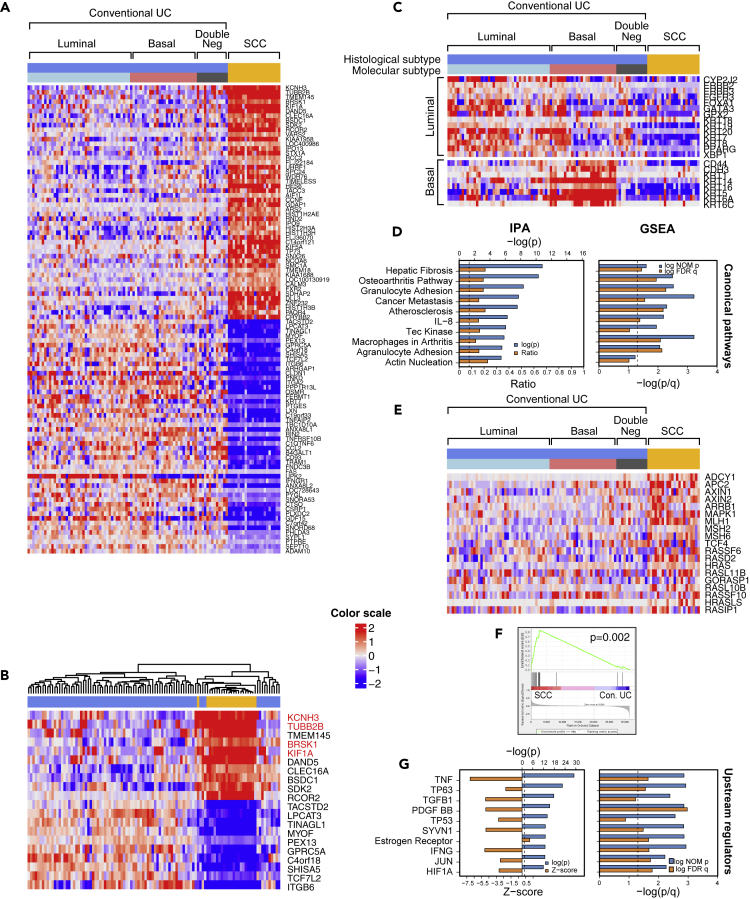
Figure 2.
Whole-Genome mRNA Expression Profile of SCC and Conventional UC
(A) The top 50 upregulated and the top 50 downregulated genes in SCC (n = 22) compared with conventional UC (n = 84).
(B) Hierarchical cluster analysis of the cohort shown in (A) using the top 10 upregulated and top 10 downregulated genes identified in SCC.
(C) Intrinsic molecular subtypes identified by the expression of luminal and basal markers in conventional UC (n = 84) and SCC (n = 22).
(D) The top 10 canonical pathways dysregulated in SCC, as revealed by Ingenuity Pathway Analysis (IPA) and Gene Set Enrichment Analysis (GSEA).
(E) Expression patterns of cancer metastasis pathway genes in molecular subtypes of conventional UC and SCC.
(F) GSEA of metastasis-related genes in SCC compared with conventional UC. A p value <0.05 was considered statistically significant. A p value < 0.05 was considered statistically significant.
(G) The top 10 upstream regulators altered in SCC, as revealed by IPA and GSEA. Genes highlighted in red are involved in neural differentiation.