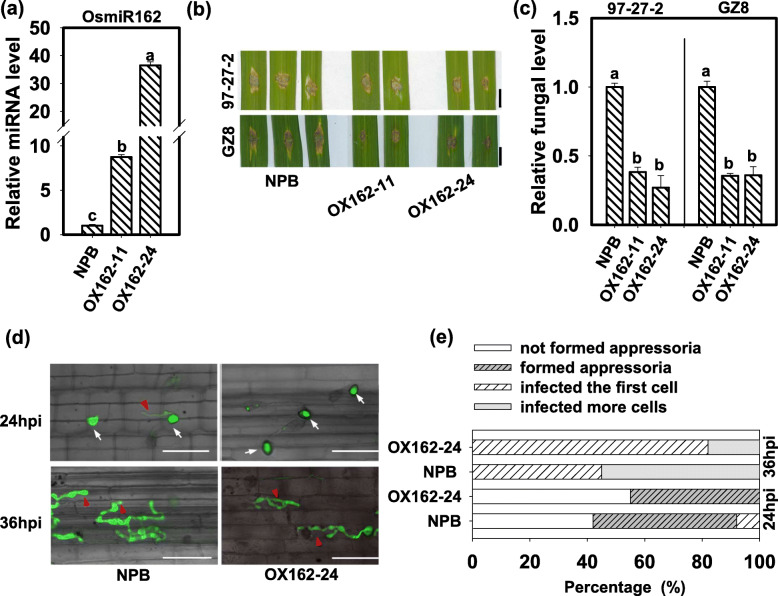
Fig. 2.
Overexpression of Osa-miR162a enhances rice resistance to Magnaporthe oryzae. a Accumulation of Osa-miR162a in transgenic lines harboring 35S: Osa-miR162a (OX162). Total RNA was used to carry out reverse-transcription with an Osa-miR162 specific stem-loop RT primers (Additional file 4: Table S1), and the RT product was subsequently used as a template for quantitative polymerase chain reaction (q-PCR) to detect the amounts of Osa-miR162. snRNA U6 served as an internal reference. b Blast disease phenotypes on leaves at 5 days post-inoculation of M. oryzae strain GZ8 and 97–27-2, respectively. Bar = 5 mm. c Relative fungal biomass of GZ8 and 97–27-2 on NPB and OX162. The relative fungal biomass was measured by using the ratio of DNA level of M. oryzae Pot2 gene against the rice genomic ubiquitin DNA level. d Invasion process of M. oryzae strain GZ8 at 24 and 36 h post-inoculation (hpi) on sheath cells of indicated lines. Bars = 25 μm. The white arrows indicate appressoria formed from conidia, and the red arrowheads indicate invasive hypha in rice sheath cells. e Quantification analysis on infection process. Over 200 conidia in each line were analyzed. For (a) and (c), Error bars indicate SD (n = 3). Different letters above the bars indicate significant differences (P < 0.05) as determined by One-way ANOVA analysis. Similar results were obtained in at least two independent experiments