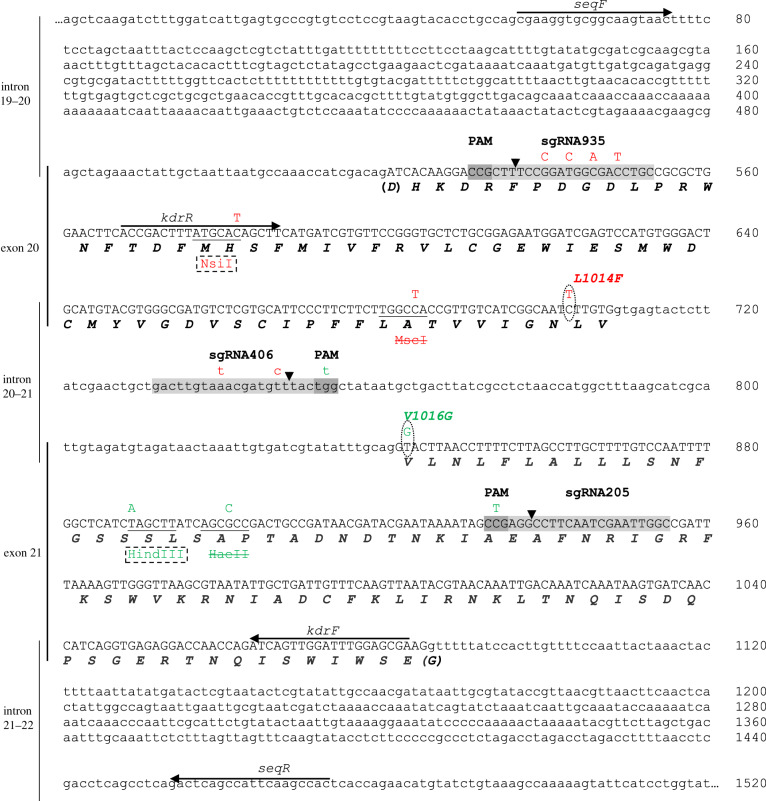
Figure 1.
CRISPR/Cas9 strategies for the generation of genome modified flies bearing mutations L1014F and V1016G. Nucleotide and deduced amino acid sequence of a 1520 bp fragment of para, encompassing exons 20 and 21 that contain positions 1014 and 1016, respectively (M. domestica numbering) of the Drosophila melanogaster amino acid sequence. Light grey areas indicate the CRISPR/Cas9 targets selected (sgRNA935, sgRNA406 and sgRNA2015), while dark grey areas indicate the corresponding PAM (-NGG) triplets. Vertical arrows denote break points for CRISPR/Cas9-induced double-stranded breaks. Red lettering indicates the differences introduced in exon 20 for the generation of L1014F, while green lettering indicates the differences introduced in exon 21 for the generation of V1016G. Ovals mark non-synonymous differences between the target (wild-type) and donor (genome modified) sequences. Synonymous mutations incorporated for diagnostic purposes, as well as to avoid cleavage of the donor plasmid by the CRISPR/Cas9 machinery, are shown above the nucleotide sequence. Restriction sites abolished because of the genome modification are shown with strikethrough letters and the corresponding sequence is underlined. Restriction sites introduced because of the genome modification are shown in dashed boxes and the corresponding sequence is also underlined. Horizontal arrows indicate the positions of primer pairs kdrF/kdrR and seqF/seqR (electronic supplementary material, table S2) used for sequencing of the genome modified alleles. (Online version in colour.)