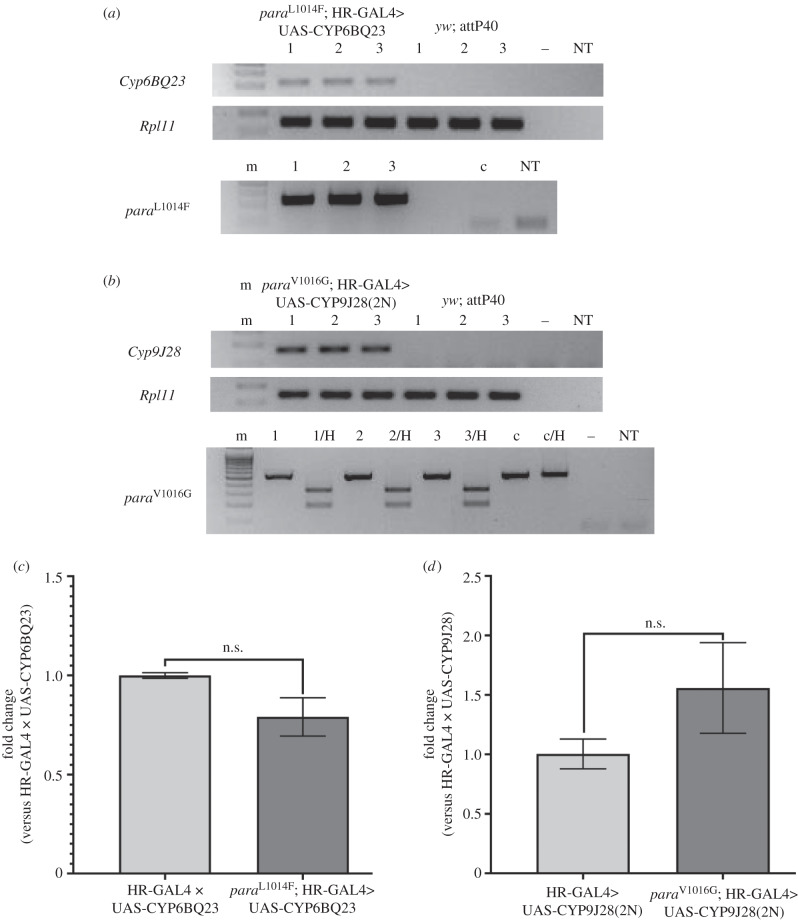
Figure 2.
P450 overexpression in the flies bearing the kdr mutations. (a) (top) Cyp6bg23 expression is confirmed by reverse transcription and PCR amplification of cDNAs. Lanes paraL1014F;HR-GAL4>UAS-CYP6BQ23 (1–3) indicate three biological replicates of the flies tested for the overexpression of the transgene. Lanes yw; attP40 (1–3) indicate the three biological replicates of the control line. The same cDNAs were used to amplify the housekeeping gene rpl11 as a reference gene. —: no reverse transcription control (to monitor for genomic DNA contamination); NT: no template control. (bottom): The presence of L1014F mutation in the same flies is tested by PCR of genomic DNA with allele-specific primers. c: yw; attP40 negative control DNA. (b) (top) Cyp9j28 expression is similarly confirmed. Lanes paraV1016G;HR-GAL4>UAS-CYP9J28(2N) (1–3) indicate three biological replicates of the flies tested for the overexpression of the transgene, while lanes yw; attP40 (1–3) indicate the three biological replicates of the control line. The same cDNAs were used to amplify the housekeeping gene rpl11 as a reference gene. —: no reverse transcription; NT: no template. (bottom): The presence of V1016G mutation in the same flies is tested by PCR of genomic DNA with generic primers and subsequent digestion of the product with HindIII (/H). c: yw; attP40 negative control DNA. (c,d) qRT-PCR for evaluation of P450 expression levels in different strains. The Ct values of strains expressing CYP6BQ23 (c) and CYP9J28 (d) were calculated in the absence or presence of the relevant para mutations. No significant difference in expression was observed (p = 0.0618 for CYP6BQ23, p = 0.1161 for CYP9J28).