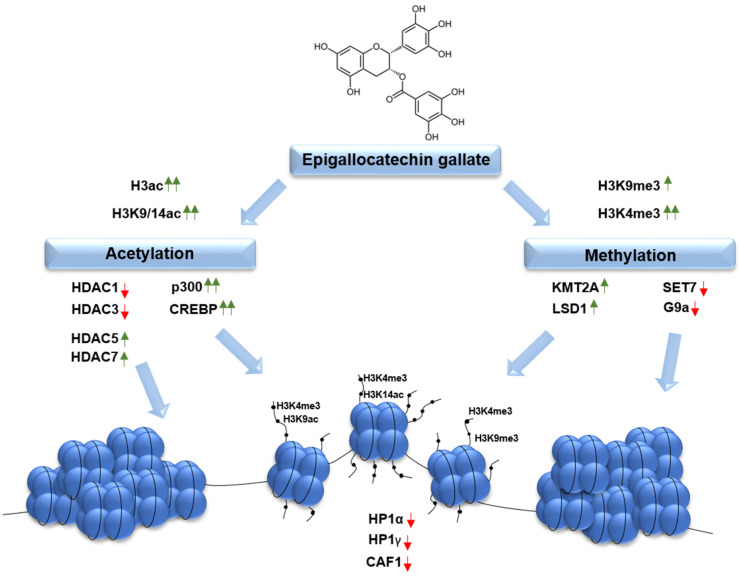
Scheme 1.
Summary of the epigenetics effects exerted by epigallocatchin-3-gallate, visualizing its potential for modification of chromatin architecture via changes in acetylation and methylation of histone proteins and DNA methylation pattern. The scheme does not include the potential crosstalk between the epigenetic modifications, however, chromatin architecture can be modified not only by the direct effect of EGCG on histone marks but also indirectly as a result of inhibition of DNMTs activity by the catchin. (HP1—heterochromatin protein 1; CAF1A—nucleosome assembly factor, subunit p150; HDAC—histone deacetylase; p300—histone acetyltransferase; CREBP—CREB-binding protein; LSD1—lysine specific demethylase 1; KMT2A—lysine methyltransferase 2A, G9a—lysine methyltransferase; CpG—or CpG island, regions of DNA rich in a cytosine followed by a guanine nucleotide).