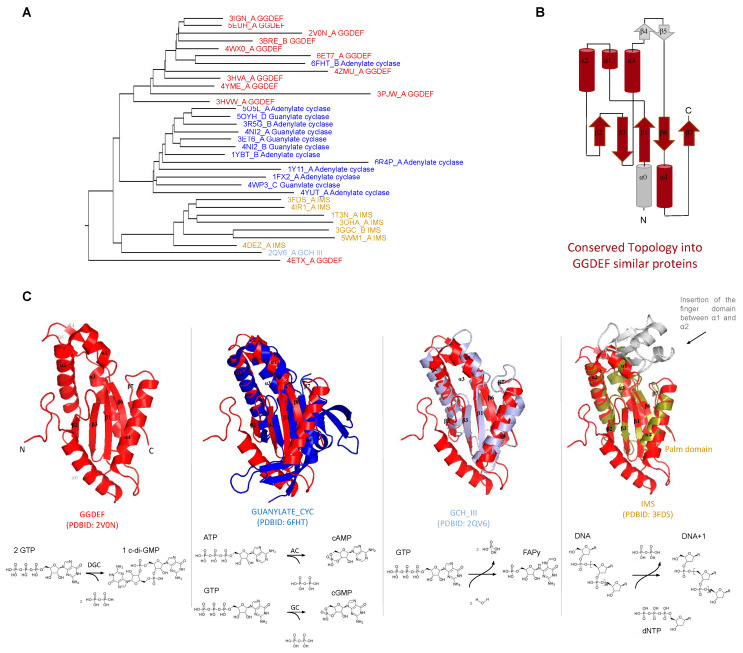
Figure 1.
Structural similarities between GGDEF (GG[D/E][E/D]F conserved sequence motif) domains and adenylate/guanylate cyclase, GTP (guanosine triphosphate) cyclohydrolase III and RRM-like palm domain of DNA polymerases. (A) dendrogram showing structures similar to GGDEF domain made with the Dali server [79] (query: PleD PDBID: 2V0N). Each domain is colored with different colors and the PDBID_chain and the Pfam name are shown for each branch. The conserved fold found in most of these structures is shown in brown, panel (B), overlaid on the GGDEF domain of PleD topology. (C) structural superposition of GGDEF domain of PleD (PDBID: 2VON) with the other domains shown in the dendrogram (panel A), using the same colors to represent each domain. At the bottom of each structural alignment, the domain’s name and the PDBID code is shown, as well as the chemical reaction performed. DGC: diguanylate cyclase; AC: adenylate cyclase, GC: guanylate cyclase, and FAPy: 2-amino-5-formylamino-6-ribosylamino-4(3H)-pyrimidinone 5‘-phosphate.