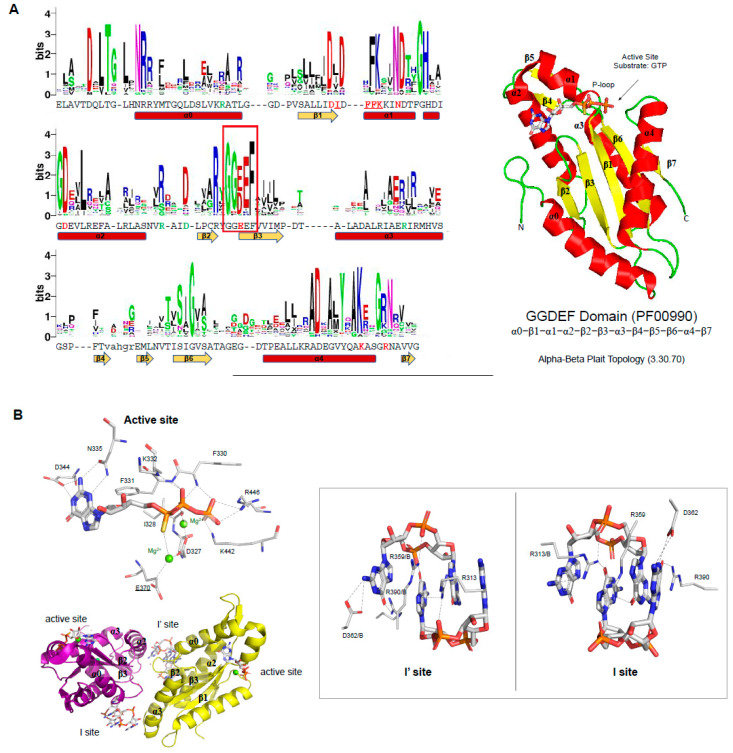
Figure 3.
Conserved sequences within GGDEF members and their active and I-sites pockets. (A) residue frequency in GGDEF domains. Using the Dali server [79], 23 sequences of GGDEF domain structures were used to create a multiple sequence alignment, and the sequence logo was created with the WebLogo server [90]. The sequence shown below the logo and the secondary structure elements belong to PleD of Caulobacter vibrioides (PDBID: 2V0N). Residues colored in red are involved in ligand or magnesium binding (for underlined residues, only the main chain is involved) and those colored in green are located in the I-sites. The GGDEF motif is placed in a red box. On the right, the structure of the GGDEF domain of PleD is shown as a cartoon. The topology of GGDEF is shown below the structure, and the CATH topology name and code are also shown [91]; (B) interaction network between the GGDEF domain of PleD binding pocket with the substrate, GTP. In the bottom, the PleD structure in the inactive conformation is shown, in which the two inhibitory sites are shown (I-site and I’-site). On the right, it is shown in more detail the residues involved in the (c-di-GMP)2 interactions at the inhibitory sites. Gray dotted lines represent hydrogen bonds. The magnesium ions are colored in green. GTP and the protein residues involved in its binding are shown as sticks. Carbons are colored white, oxygens are red, nitrogen atoms are blue, and phosphorous atoms are orange.