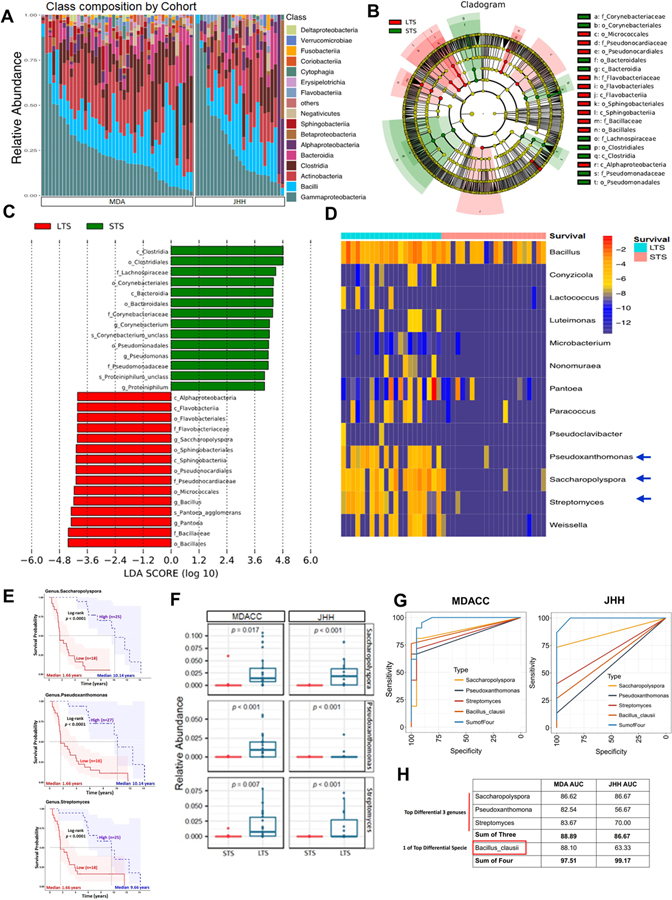
Figure 2.
Tumor microbiome communities are significantly different between LTS and STS.
(A) Bar plots of the class taxonomic levels in MDA and JHH cohorts of PDAC patients. Relative abundance is plotted for each tumor.
(B) Taxonomic Cladogram from LEfSe, depicting taxonomic association from between microbiome communities from LTS and STS PDAC patients. Each node represents a specific taxonomic type. Yellow nodes denote the taxonomic features that are not significantly differentiated between LTS and STS. Red nodes denote the taxonomic types with more abundance in LTS than in STS, while the green nodes represent the taxonomic types more abundant in STS.
(C) LDA score computed from features differentially abundant between LTS and STS. The criteria for feature selection is Log LDA Score > 4.
(D) Heatmap of selected most differentially abundant features at the genus level. Highlighting three taxa enriched in LTS. The blue color represents less abundant, lighter yellow color represents intermediate abundance and red represents the most abundant. Highlighting three taxa enriched in LTS.
(E) Kaplan-Meier estimates for survival probability based on the abundance levels of microbes enriched at Genus level in LTS. Right plot, Saccharopolyspora, middle plot, Pseudoxanthomonas and left plot, Streptomyces (p < 0.0001).
(F) Plots of differentially abundant genus significantly enriched in both MDA and JHH LTS patients. FDR adjusted p-values from negative binomial test p-value.
(G) ROC analysis of Taxa relative abundance as predictive of LTS status. The top 3 differential bacteria (Genus) identified and Baccilus Clausii (One of top species) were tested individually and in aggregate in the MDA Discovery Cohort (Left panel) were then validated in the JHH Validation Cohort (Right panel).
(H) Table depicting AUC of bacteria tested in Fig2G for both MDA and JHH cohorts.