Abstract
Purpose
It has been suggested that the biologic characteristics of breast cancer may differ among different geographic or ethnic populations. Indeed, triple-negative breast cancer (TNBC), the most lethal breast cancer subgroup, has been reported to occur at a higher incidence in Japan than in the United States. However, most genomic studies of these tumors are from Western countries, and the genomic landscape of TNBC in an Asian population has not been thoroughly investigated. Here, we sought to elucidate the geographic and ethnic diversity of breast cancer by examining actionable driver alterations in TNBC tumors from Japanese patients and comparing them with The Cancer Genome Atlas (TCGA) database, which gathers data primarily from non-Asian patients.
Materials and Methods
We performed comprehensive genomic profiling, including an analysis of 435 known cancer genes, among Japanese patients with TNBC (n = 53) and compared the results with independent data obtained from TCGA (n = 123).
Results
Driver alterations were identified in 51 (96%) of 53 Japanese patients. Although the overall alteration spectrum among Japanese patients was similar to that of TCGA, we found significant differences in the frequencies of alterations in MYC and PTK2. We identified three patients (5.7%) with a high tumor mutational burden, although no microsatellite instability was observed in any of the Japanese patients. Importantly, pathway analysis revealed that 66.0% (35 of 53) of Japanese patients, as well as 66.7% (82 of 123) of TCGA cohort, had alterations in at least one actionable gene targetable by US Food and Drug Administration–approved drug.
Conclusion
Our study identified actionable driver alterations in Japanese patients with TNBC, revealing new opportunities for targeted therapies in Asian patients.
INTRODUCTION
Although breast cancer is one of the most frequently diagnosed cancers among women in the United States and Japan,1,2 its biologic characteristics may differ among different geographic or ethnic populations. Indeed, breast cancer incidence, peak age of highest incidence, and mortality are different between women in the United States and Japan.3 Importantly, triple-negative breast cancer (TNBC), the most lethal subgroup of breast cancer and characterized as estrogen receptor negative and progesterone receptor negative with no overexpression of human epidermal growth factor receptor 2/neu,4 is known to be more prevalent in Japan than in the United States.5,6
Large-scale genomic studies based on next-generation sequencing (NGS), such as The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium, have revealed important driver gene alterations in many types of solid tumors,7-14 which have led to new targeted therapeutic strategies for precision cancer medicine.15-19 These studies have also highlighted the geographic and ethnic diversity in the cancer genome.20,21 However, most NGS-based studies have been conducted in Western countries, where the Asian population is a minority, and the resulting genomic information may not truly represent this population.8 For instance, only 5.5% of participants have identified themselves as Asians in the TCGA cohort. Therefore, it is of interest to clarify whether there is a genetic difference in TNBC between Asian and Western patients. In our study, we hypothesized that any differences in genetic driver events may reflect differences in the biologic etiology of TNBC between Asian and Western patients.
MATERIALS AND METHODS
Patients
The Japanese cohort comprised a total of 53 patients diagnosed with stage I to IV TNBC according to the American Joint Committee on Cancer (seventh edition)22 between 2009 and 2016 at Niigata University Medical and Dental Hospital, Niigata Cancer Center Hospital, Keio University Hospital, Gifu University Hospital, Showa University Hospital, or St Luke’s International Hospital. Collection and use of all specimens in this study were approved by the institutional review board of each institution. Informed consent was obtained from all participants. TNBC tumors in both the Japanese cohort and the TCGA database were defined according to previous reports.23,24
Sequencing Library Preparation
Archival tissue in the formalin-fixed, paraffin-embedded (FFPE) tumor obtained during routine biopsy or resection of primary tumor was used for analysis. An independent pathologist evaluated the tumor content on hematoxylin and eosin–stained slides for each study sample to ensure > 50% tumor content was present. All sample preparation, comprehensive genomic sequencing, and analytics were performed in a Clinical Laboratory Improvement Amendments/ College of American Pathologists–accredited laboratory (KEW, Cambridge, MA) as described previously.19
Comprehensive Genomic Sequencing
FFPE genomic DNA (50 to 150 ng) was used to construct libraries that were enriched for 435 genes using CANCERPLEX (KEW) as described previously.19 The term gene alterations is used when there are genetic aberrations such as amplifications in addition to mutations. To assess the somatic status of alterations in tumor-only settings, we used a filtering strategy similar to one recently published25 with minor differences. Detailed methodologies including clustering are described in the Data Supplement.
Genomic and Clinicopathologic Data for the TCGA TNBC Cohort
Genomic and clinicopathologic data from a total of 123 tumor samples of TNBC in the TCGA database were downloaded from cBioPortal (www.cbioportal.org). BRCA alteration data for the TCGA TNBC samples were downloaded from the Broad GDAC Firehose Web site (https://gdac.broadinstitute.org/). As in the 435-gene panel bioinformatics pipeline, silent mutations that did not alter the amino acid sequence were removed from the data set. To compare the mutational burden of the 435-gene panel with that of the TCGA whole-exome sequencing (WES) data, the data set of single-nucleotide polymorphisms was downsampled to the 435 genes in the panel, and the panel mutation rate was determined, calculated as mutations per megabase, as described previously.19
RESULTS
Genetic Alterations in Japanese Patients With TNBC
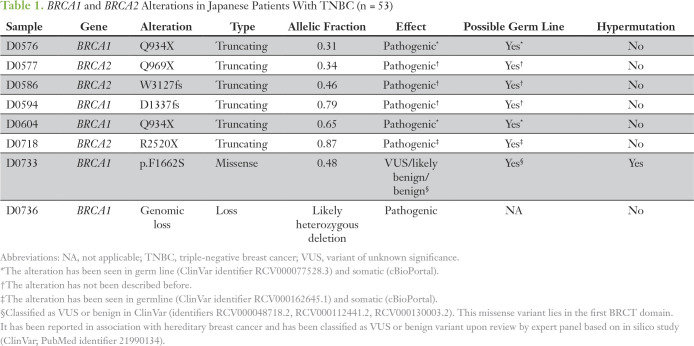
The clinicopathologic characteristics of the patients with TNBC in the Japanese and TCGA cohorts are summarized in the Data Supplement. The Japanese cohort included more patients with M1 disease (11%) compared with the TCGA cohort (1%; P < .01), which seemed to affect the difference in stage among the two cohorts (Data Supplement). After performing comprehensive genomic sequencing on all Japanese TNBC samples, we found at least one alteration in 82 of 435 cancer-associated genes (Fig 1; Data Supplement). Although a total of 186 alterations were found in the 53 patients, there were only three frequently altered genes (seen in > 10% of patients): TP53, PIK3CA, and PTEN (Fig 1). Alterations of BRCA1 and BRCA2 presented in 9.4% and 5.6% of patients, respectively, totaling 15.1% of the population (Fig 1).
Fig 1.
Comparison of genetic alterations in triple-negative breast cancer between the Japanese and The Cancer Genoma Atlas (TCGA) cohorts. Alteration frequencies of cancer-associated genes in Japanese patients detected by a 435-gene panel, and alteration frequencies for the same gene set in the TCGA cohort. Statistical significance was determined using Fisher’s exact test. (*) P < .05 for alteration frequencies between Japanese patients and the TCGA cohort.
The Japanese cohort included 35 patients who received neoadjuvant chemotherapy. Among them, biopsy specimens of primary tumors before neoadjuvant chemotherapy were used for analysis in 11 patients, and surgical specimens of remnant primary tumors after neoadjuvant chemotherapy were used in 24 patients (Data Supplement). We compared gene alterations in chemotherapy-naïve tissue samples (n = 29) and chemotherapy-affected tissue samples (n = 24) and found that the gene alteration frequencies, including those for TP53, PIK3CA, and PTEN, were comparable between the two groups (Data Supplement). We also extracted gene alteration data from 123 cases in the TCGA TNBC cohort for the same set of genes as in Japanese patients to compare our results with TCGA data (Fig 1; Data Supplement). Of note, there were two distinct categories in the TCGA cohort: TNBC based on pathologic subtyping and basal-like breast cancer based on PAM50 analysis, with some overlap between the two groups. Interestingly, the TP53 alteration rate in TNBC (65 [52.8%] of 123) was relatively low in the TCGA cohort, although the rate in basal-like breast cancer (73 [78.5%] of 93) was much higher in the same cohort (Data Supplement). Generally, the gene alteration rates were comparable between Japanese patients and the TCGA cohort; however, the alteration frequencies in two genes (MYC and PTK2) were significantly different between the two cohorts (Fig 1; Data Supplement).
Genomic Alterations in Cancer Signaling Pathways
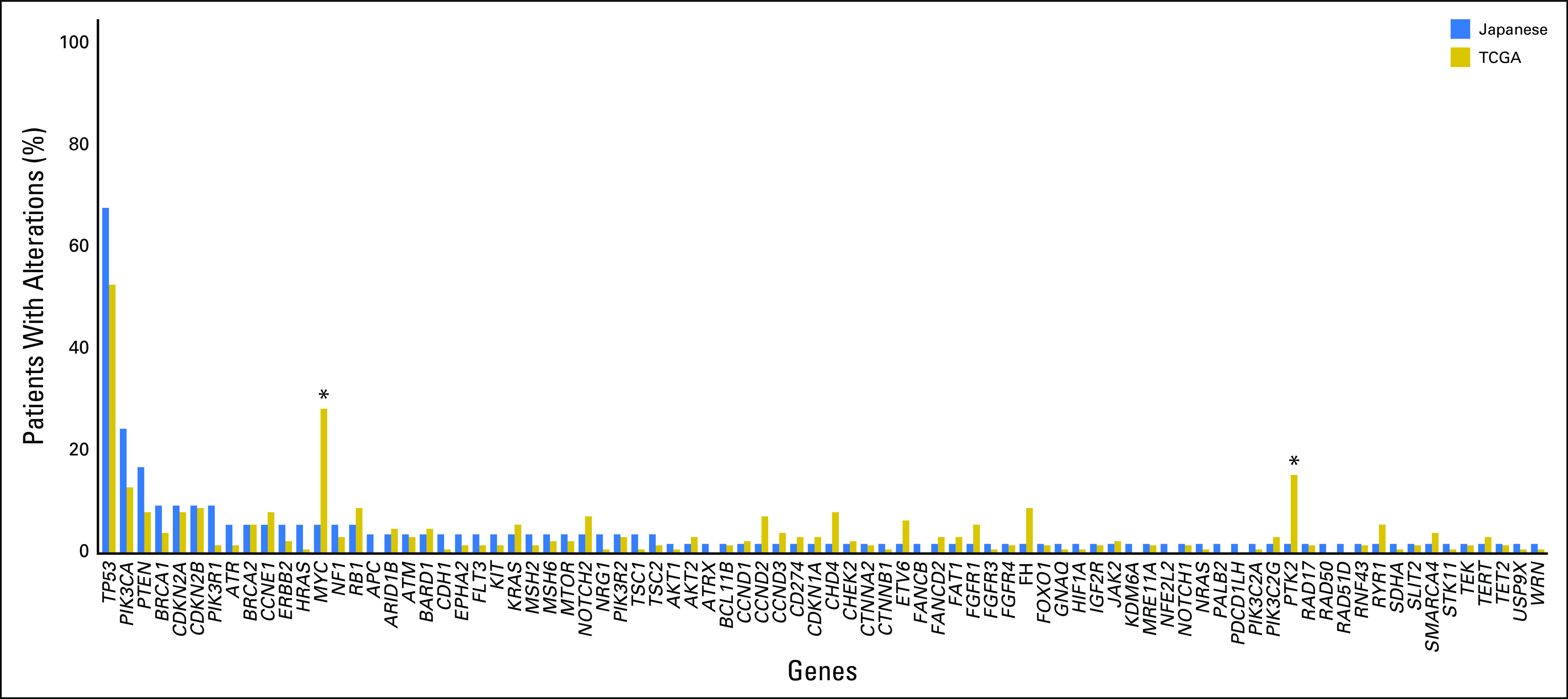
We next assessed genomic alterations in major oncogenic pathways involving phosphatidylinositol 3-kinase, the cell cycle, DNA double-strand break (DSB) repair, ERBB2/KRAS, and β-catenin/WNT signaling and determined whether there were differences between the two cohorts (Figs 2A to 2J). Overall, the gene alteration spectrum of the Japanese cohort was similar to that of the TCGA cohort (Figs 2A to 2J). However, there were some exceptions, such as significantly more amplification of MYC in the TCGA cohort (28%) than in Japanese patients (6%; P < .01; Fig 2).
Fig 2.
Genetic alterations across common oncogenic pathways in triple-negative breast cancer. (A-E) Japanese patients (n = 53) and (F-J) The Cancer Genome Atlas (TCGA) cohort (n = 123) were evaluated for gene alterations in key cancer pathways: (A, F) phosphatidylinositol 3-kinase (PI3K; 45% and 25% of patients, respectively), (B, G) cell cycle (26% and 29% of patients, respectively), (C, H) DNA double-strand break (DSB) repair (75% and 58% of patients, respectively), ERBB2/KRAS (17% and 11% of patients, respectively), and (E, J) β-catenin/WNT (11% and 29% of patients, respectively). Altered cases are defined as the total number of unique samples with a genetic alteration in each pathway. (K-O) Percentage of patients with a variation for each given gene: (K) PI3K, (L) cell cycle, (M) DNA DSB repair, (N) ERBB2/KRAS, and (O) β-catenin/WNT. Statistical significance was determined using Fisher’s exact test. (*) P < .05. (†) P < .01.
We found that eight patients (15%) showed BRCA1/2 alterations, including five BRCA1 and three BRCA2 alterations (Figs 2A to 2J). There was no difference in the frequency of BRCA1/2 alterations between the Japanese and TCGA cohorts (Figs 2K to 2O). We further analyzed the BRCA1/2 alterations in the Japanese cohort (Table 1). On the basis of the alteration site, seven of eight alterations were considered to be pathogenic alterations (Table 1). Moreover, bioinformatic analysis revealed that seven of eight alterations were possible germ line alterations, based on the allelic fraction and alteration site compared with a database, such as ClinVar (Table 1). We did not confirm the germ line alteration using patients’ normal tissue.
Table 1.
BRCA1 and BRCA2 Alterations in Japanese Patients With TNBC (n = 53)
Tumor Mutational Burden and Microsatellite Instability in Patients With TNBC
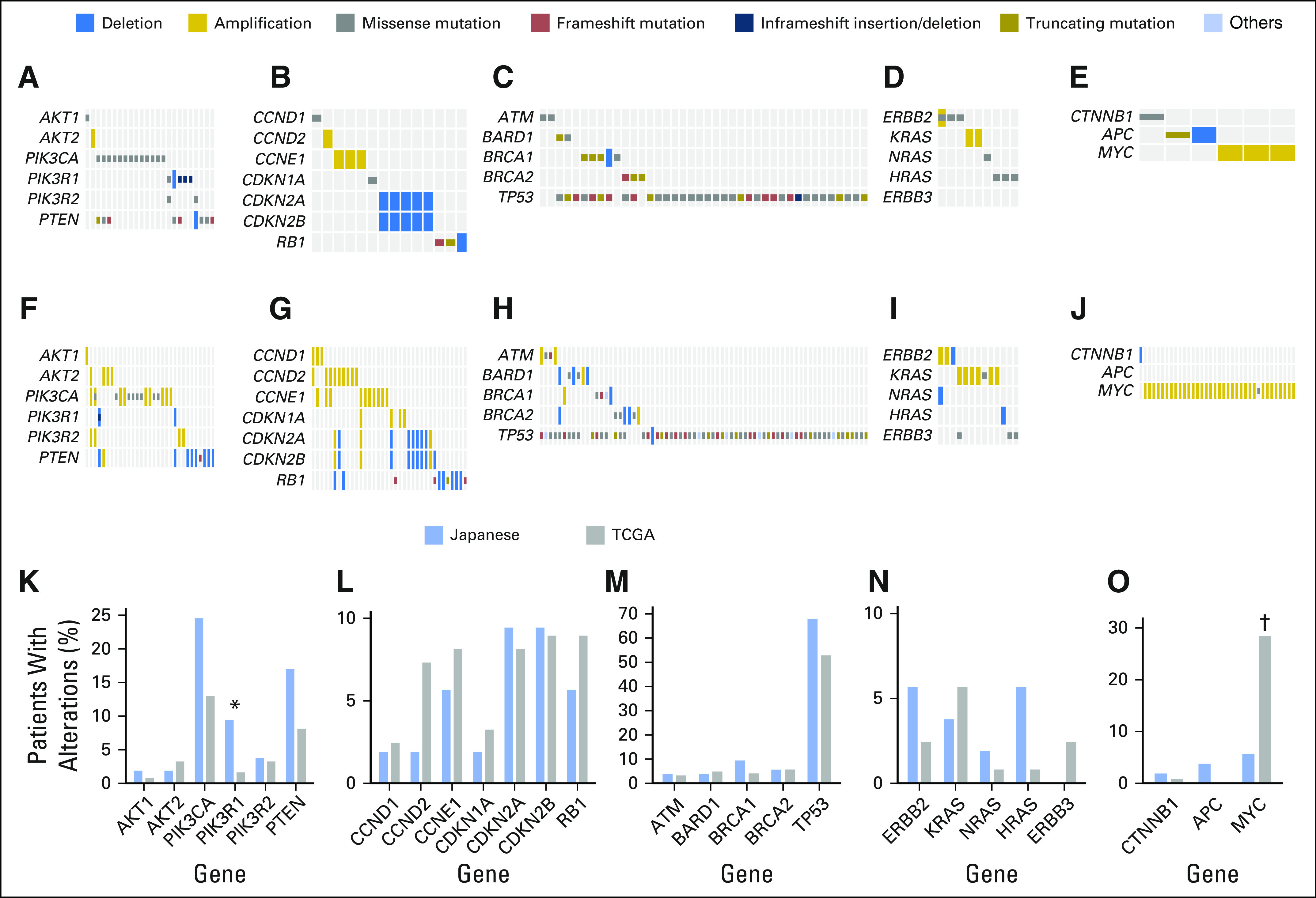
A high tumor mutational burden (TMB-H) tumor is defined as a tumor with a high rate of somatic mutation. TMB-H tumors were recently correlated with the generation of neoantigens and clinical response to immunotherapy drugs.7,26-29 The median mutation rate for Japanese patients was 11.5 mutations/Mb (range, 3.9 to 56.2 mutations/Mb). We identified three patients (5.7%) as TMB-H (≥ 20 mutations/Mb of sequenced DNA). Of note, one of the three TMB-H patients showed BRCA1 alteration (Table 1). This classification was based on our previous studies of colorectal and gastric cancers, which included a number of patients with microsatellite instability, although no microsatellite instability was observed in any of the 53 Japanese patients with TNBC (Fig 3A). To compare the mutational burden in Japanese patients with that in the TCGA cohort, we downsampled the TCGA WES data (n = 123 tumors) to the subset of 435 genes in the panel test platform. This analysis not only accurately identified TMB-H tumors but also showed strong correlation in mutation rates between the 435-gene panel and WES (R2 = 0.9203; Fig 3B). The average mutation rate detected by the 435-gene panel (8.6 mutations/Mb) was higher than that detected by WES (1.7 mutations/Mb), reflecting the fact that the panel content was in part selected to include genes more frequently mutated in cancer, as reported previously.19 The mutational burden of the TCGA TNBC cohort reanalyzed in silico revealed a comparable mutational burden between the Japanese cohort and the TCGA cohort, in which six (4.9%) were determined as TMB-H (Fig 3).
Fig 3.
Genetic alterations related to potential targeted therapies. (A) Mutation rate and microsatellite instability (MSI) in patients with triple-negative breast cancer in Japan (n = 53); three (5.7%) of 53 hypermutated. (B) Mutation rate data from whole-exome sequencing (WES) for The Cancer Genome Atlas cohort (n = 123) were downsampled to the content of the 435-gene panel platform; six (4.9%) of 123 hypermutated. Inset: correlation between mutation rates determined by 435-gene panel and WES.
Clustering Analysis for Gene Alterations in Patients With TNBC
Next, we performed hierarchic clustering of alterations in a subset of genes associated with an US Food and Drug Administration (FDA) –approved therapy (on or off label; n = 68 genes) across all 176 TNBC samples (Japanese, n = 53 plus TCGA, n = 123) to further assess the characteristics of genomic alterations in TNBC (Fig 4). We determined that all patients could be classified into 10 typical clusters (Fig 4B). Cluster one (n = 83; 47.2%), in which no common actionable alterations were seen, was the largest of the 10 identified clusters, and cluster nine (n = 23; 13.1%), in which PIK3CA alterations were common, was the second largest cluster (Fig 4B). Cluster seven (n = 16) was characterized by PTEN alterations, cluster eight (n = 12) was characterized by CDKN2A and CDKN2B alterations, and clusters six (n = 6) and three (n = 6) were characterized by BRCA1 and BRCA2 alterations, respectively. Moreover, cluster two (n = 7) contained FGFR1 alterations, cluster four (n = 7) contained DDR2 alterations, and cluster five (n = 10) contained CCND2/3 alterations. Finally, cluster 10 (n = 6) was characterized by multiple gene alterations, including PIK3CA, PTEN, and CDKN2A/B. In summary, the large cluster one, without any distinct alterations, and other clusters with typical alternated genes highlighted the great diversity of alterations underlying TNBC, which consequently constrained our clustering analysis designed to match genomic alterations with FDA-approved drugs.
Fig 4.
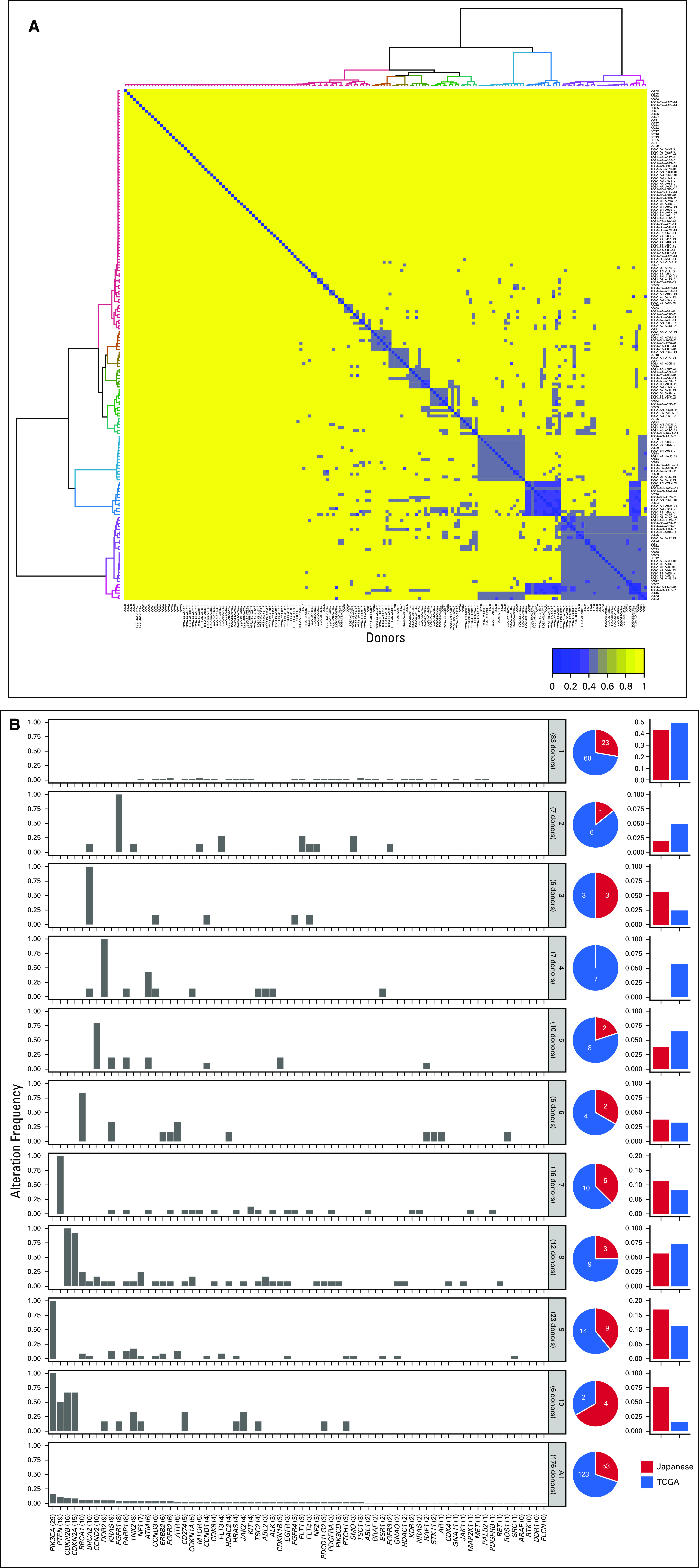
Cluster of 68-gene coalteration patterns. (A) A cluster analysis was performed on 176 cases of triple-negative breast cancer (TNBC; 53 tumors from Japanese patients and 123 tumors from The Cancer Genome Atlas [TCGA] cohort) by using Euclidean distance and Ward’s clustering method (distance based on common mutated genes are colored blue to yellow). The 68 genes associated with a US Food and Drug Administration–approved drug (on or off label) were used for clustering. (B) Comutated gene patterns of a 68-gene set. The percentage of Japanese patients and the percentage of the TCGA cohort in each cluster are shown in the pie graphs displayed on the right side. The alteration rate in each cluster is shown as a bar graph in the far right.
Actionable Alterations for Potential Treatments
Because the cluster analysis did not effectively identify potential treatment strategies on the basis of individual genomic alterations, we therefore assessed the possibility of using pathways associated with FDA-approved drugs to identify targeted therapies for patients with TNBC. We determined that 29 of 82 mutated genes were actionable with FDA-approved drugs (on or off label). Importantly, 35 patients (66.0%) had at least one potentially actionable alteration associated with FDA-approved drugs, whereas 18 patients had no actionable alterations with those drugs (Fig 5). Interestingly, we similarly analyzed the TCGA cohort and found that, as with the Japanese cohort, 66.7% of cases had at least one potentially actionable alteration associated with FDA-approved drugs.
Fig 5.
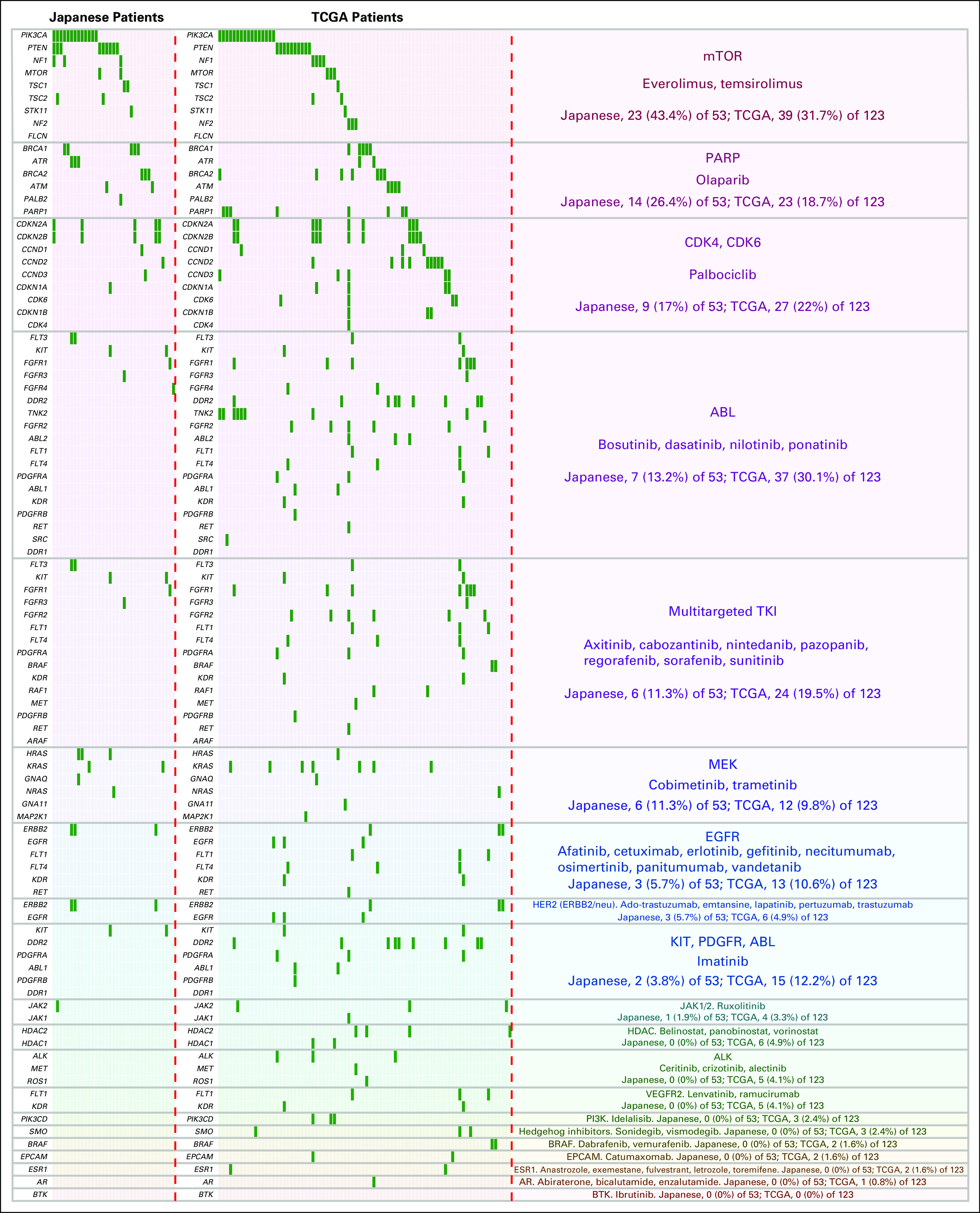
Genetic alterations in Japanese patients with triple-negative breast cancer and patients in The Cancer Genome Atlas (TCGA) cohort potentially targetable by US Food and Drug Administration (FDA) –approved cancer therapies. The genes associated with an FDA-approved drug (on or off label) are listed, and each individual genetic alteration is indicated as a green bar. The FDA-approved cancer therapies are shown in the right column. The alteration rates in the Japanese population (n = 53; alterations, 35 [66%] of 53) and TCGA cohort (n = 123; alterations, 82 [66.7%] of 123) are also shown in the right column. AR, androgen receptor; BTK, Bruton tyrosine kinase; EGFR, epidermal growth factor receptor; EPCAM, epithelial cell adhesion molecule; HDAC, histone deacetylase; mTOR, mammalian target of rapamycin; PARP, poly (ADP-ribose) polymerase; PDGFR, platelet-derived growth factor receptor; PI3K, phosphatidylinositol 3-kinase; TKI, tyrosine kinase inhibitor; VEGFR2, vascular endothelial growth factor receptor 2.
For Japanese patients, genes associated with mammalian target of rapamycin (mTOR), poly (ADP-ribose) polymerase (PARP), and CDK4/6 inhibitors showed the three highest alteration rates (Fig 5). Twenty-three patients (43.4%) showed actionable alterations that could be targeted by mTOR inhibitors, whereas 31.7% of TCGA cases showed actionable alterations in this pathway. Importantly, 26.4% of Japanese patients and 18.7% of TCGA cases had actionable alterations related to the DSB repair pathway, which could be potentially treated by PARP inhibition. Seventeen percent of Japanese patients and 22% of TCGA cases had alterations in actionable genes related to the cell-cycle pathway, which could be treated by CDK4/6 inhibition. Moreover, other FDA-approved drugs, including multitargeted tyrosine kinase inhibitors, MEK inhibitors, anti–human epidermal growth factor receptor 2 therapies, and JAK inhibitors, were indicated as potential targeted therapies for Japanese patients with TNBC, with even more drugs suggested for TCGA cases (Fig 5). Of note, Oncoprint analysis showed coalterations in many cases of TNBC in both cohorts, which may be associated with resistance to the targeted therapies (Fig 5). Additional investigation, including clinical outcomes, is needed to reveal the efficacy of these therapies for patients with TNBC.
DISCUSSION
We found significant differences in the frequencies of gene alterations, including MYC amplification, between Japanese patients with TNBC and the TCGA TNBC cohort. However, the overall alteration spectrum of the Japanese patients was similar to that of the TCGA cohort.8 Importantly, 66% of Japanese patients, as well as 67% of TCGA cases, had at least one genomic alteration in actionable genes, providing potentially new treatment strategies.
We demonstrated significant differences in the frequencies of alterations in two genes: PTK2 and MYC. It has been previously reported that MYC is one of the most important driver genes, and there are known ethnic differences in MYC amplification frequency.30 Although we found a MYC amplification rate of 5.7% in our cohort, high incidence (30% to 35%) of MYC amplification has been repeatedly reported from US groups, similar to TCGA rates.31-33 A unique patient population comprises the cohort described in our study, which could be useful given the sparse genomic data from Asian TNBC populations.
We analyzed genomic alterations in FFPE tumor tissue from a cohort of Japanese patients with TNBC by comprehensive NGS-based sequencing of a panel of 435 cancer-relevant genes, at an average 500× depth. In contrast, TCGA data were obtained by WES of fresh frozen tumor tissue, with a mean coverage of 97.6× depth. FFPE samples are more commonly available than fresh frozen samples. Method of preservation is critical for DNA analysis using FFPE tissue samples, as we reported previously.34 The quality of all DNA samples used in this study was confirmed to be adequate for accurate NGS analysis by strict quality control checks. Indeed, most of the genes altered in both the Japanese and TCGA cohorts showed similar frequencies, consistent with previous reports, despite the different sequencing techniques used.
Previous comprehensive genomic profiling studies have reported that TNBC is a heterogeneous disease involving diverse genomic alterations.8,35,36 Our results also demonstrated the heterogeneity of TNBC, with only three genes mutated in > 10% of patients and most of the genetic alterations differing from one another. Moreover, clustering analysis of patients with TNBC on the basis of genetic alterations also emphasized the heterogeneous nature of TNBC. Because of this heterogeneity, it was difficult to find common actionable driver alterations for therapies, and our results demonstrated that a more comprehensive approach, such as the one used in this study, will be required to find actionable alterations in each patient with TNBC.
To explore the clinical utility of targeted sequencing with a large panel for TNBC, we identified actionable genes that can be treated by FDA-approved drugs and found that 35 Japanese patients (66%) had at least one such actionable gene alteration. We identified several signaling pathways for molecular targeted therapies, including the PI3K pathway for mTOR inhibitors, the DSB repair pathway for PARP inhibitors, and the cell-cycle pathway for CDK4/6 inhibitors (Fig 5). These important pathways in Japanese TNBC are comparable to those in previous reports from Western countries,8,31,36,37 and potential targeted therapies for these pathways could be similar, regardless of ethnicity.18,38
In addition to identifying driver alterations, the clinical significance of identifying hypermutated tumors was recently demonstrated in several studies correlating mutational burden with the development of neoantigens and clinical response to immunotherapy drugs.7,26-29,39 We previously showed that a panel of 400+ genes was able to identify TMB-H tumors with sensitivity and specificity comparable to those of WES in colorectal cancer.19 Similarly, here we determined the TMB and revealed that a small proportion of patients (5.7%) were hypermutated. Although none of these patients received immune checkpoint inhibitor therapy, there is a high likelihood that these patients may have responded, given clinical data demonstrating efficacy in TMB-H patients.
In our study, we found that seven (13%) of 53 Japanese patients with TNBC had possible germ line BRCA1/2 alterations based on bioinformatic analysis of tumor DNA. Although specific genetic tests are needed to confirm the frequency of germ line BRCA1/2 alterations, this estimated frequency is in agreement with previous reports.40 Recently, germ line alteration of BRCA1/2 has attracted increasing attention because of the use of PARP inhibitors. Considering the frequency of germ line BRCA1/2 alterations, it seems that there is a certain population that would benefit from treatment with PARP inhibitors, not only in Western countries, but also in Asian countries, including Japan.
Patients were approached consecutively. However, the number of patients enrolled in this study was limited, partly because of certain misunderstandings and prejudices that prevent some Japanese patients consenting to genomic sequencing. Power analyses for all genes were not conducted before this study; however, we did verify the test power based on gene alteration rates of 53 Japanese samples. As a result, we determined that a Fisher’s exact test of significant genes MYC and PTK2 met adequate power (≥ .8). Therefore, we considered 53 as a sufficient number for the Japanese cohort, at least to the extent that the alteration rate of more than one gene was significantly different between the Japanese and TCGA cohorts.
Another limitation is that we included patients receiving neoadjuvant chemotherapy, and we cannot exclude the possibility that the neoadjuvant chemotherapy affected genomic alterations. However, when we compared the gene alterations between the chemotherapy-naïve tissue samples (n = 29) and chemotherapy-affected tissue samples (n = 24), we found that gene alteration frequencies, including those for TP53, PIK3CA, and PTEN, were comparable between the two groups (Data Supplement). Moreover, considering that we did not see major differences in genomic alterations between the Japanese and TCGA cohorts except for MYC, it is unlikely that neoadjuvant chemotherapy dramatically influenced the landscape of driver alterations. Although it is not easy yet in Japan to recruit a large number of patients with TNBC for genomic sequencing, additional studies with a greater number of patients are warranted to confirm our findings in the future.
Despite these limitations, to our knowledge, this is the largest cohort of Japanese patients with TNBC to undergo comprehensive genomic profiling to date, and our data provide important insights into the molecular heterogeneity underlying this aggressive disease from an ethnogeographic perspective. Similarly, several studies have demonstrated the heterogeneity of TNBC,5,41-43 therefore suggesting that targeting TNBC by molecular targeted therapies remains a challenge. Indeed, our results showed coalterations in many TNBC cases in both cohorts, potentially associated with resistance to targeted therapies. Although our study indicates the possibility of targeted therapies for patients with TNBC, an accumulation of clinical evidence will be required to reveal the usefulness of each therapy for this disease. Additional clinical studies are required to reveal potential treatments for patients with TNBC.
In conclusion, our study revealed actionable driver alterations in Japanese patients with TNBC that could potentially be treated with existing drugs. Although we found significant differences in the frequencies of gene alterations between Japanese and TCGA TNBC patients, which included MYC amplification, potential targeted therapies for patients with TNBC could be similar regardless of ethnicity. Our study has revealed new opportunities for the use of targeted therapies in the Asian population.
ACKNOWLEDGMENT
We thank Jennifer Ring for English editing.
Footnotes
Supported by Denka; Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research Grants No. JP15H05676 and JP15K15471, the Takeda Science Foundation, the Tsukada Medical Foundation, and the Tohoku Cancer Professional Training Promotion Plan (M. Nagahashi); JSPS Grant-in-Aid for Scientific Research Grants No. JP15H04927 and JP16K15610 (T.W.); JSPS Grant-in-Aid for Scientific Research Grant No. JP16K19888 (J.T.); JSPS Grant-in-Aid for Scientific Research Grant No. JP26700029 (S.O.); National Cancer Institute, National Institutes of Health, Grant No. R01CA160688 and Susan G. Komen Investigator Initiated Research Grant No. IIR12222224 (K.T.); and a grant from the Massachusetts Life Sciences Center (S.L.).
AUTHOR CONTRIBUTIONS
Conception and design: Masayuki Nagahashi, Kazuhiro Yoshida, Kazuaki Takabe, Toshifumi Wakai
Administrative support: Yoshifumi Shimada, Toshifumi Wakai
Provision of study material or patients: Manabu Futamura, Kazuhiro Yoshida, Hideko Yamauchi
Collection and assembly of data: Masayuki Nagahashi, Tetsu Hayashida, Yuko Kitagawa, Manabu Futamura, Kazuhiro Yoshida, Takashi Kuwayama, Seigo Nakamura, Chie Toshikawa, Hideko Yamauchi, Teruo Yamauchi, Koji Kaneko, Chizuko Kanbayashi, Nobuaki Sato, Yasuo Miyoshi, Junko Tsuchida, Masato Nakajima, Hiroshi Ichikawa, Stephen Lyle
Data analysis and interpretation: Masayuki Nagahashi, YiWei Ling, Kazuhiro Yoshida, Yoshifumi Shimada, Stephen Lyle, Kazuaki Takabe, Shujiro Okuda, Toshifumi Wakai
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Actionable Gene Alterations in an Asian Population With Triple-Negative Breast Cancer
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Masayuki Nagahashi
Research Funding: Denka (Inst)
YiWei Ling
No relationship to disclose
Tetsu Hayashida
Honoraria: AstraZeneca, Novartis, Eisai, Chugai Pharma, Kyowa Hakko Kirin, Takeda Pharmaceuticals, Daiichi Sankyo
Research Funding: Taiho Pharmaceutical (Inst), Daiichi Sankyo (Inst), Chugai Pharma (Inst), Eisai (Inst), Kyowa Hakko Kirin (Inst)
Yuko Kitagawa
Honoraria: Ethicon, Olympus, Ono Pharmaceutical, Taiho Pharmaceutical, Chugai Pharma, Nippon Kayaku, Asahi Kasei
Research Funding: Astellas Pharma (Inst), Otsuka (Inst), Kaken Pharmaceutical (Inst), Kyowa Hakko Kirin (Inst), Kowa (Inst), CSL Behring (Inst), Shionogi Pharma (Inst), Daiichi Sankyo (Inst), Taiho Pharmaceutical (Inst), Takeda Pharmaceuticals (Inst), Chugai Pharma (Inst), Tsumura (Inst), Teijin Pharma (Inst), Medtronic (Inst), Boehringer Ingelheim (Inst), Merck Serono (Inst), Novartis (Inst), Ajinomoto (Inst), Asahi Kasei (Inst), Otsuka (Inst), Kureha (Inst), Sanofi (Inst), Dainippon Sumitomo Pharma (Inst), Taisho Toyama Pharma (Inst), Nippon Kayaku (Inst), Eli Lilly (Inst), Pfizer (Inst), Yakult Honsha (Inst), GlaxoSmithKline (Inst), Medicon (Inst), EA Pharma (Inst)
Manabu Futamura
No relationship to disclose
Kazuhiro Yoshida
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Takeda Pharmaceuticals, Eli Lilly, Daiichi Sankyo, Ono Pharmaceutical, Merck Serono, EA Pharma, Johnson & Johnson, Covidien, Bayer Yakuhin, Eisai, Olympus, Terumo, Bristol-Myers Squibb Japan, Otsuka, Sanofi, Denka, Nippon Kayaku, Merck Sharp & Dohme, Yakult Honsha
Research Funding: Sanofi (Inst), Yakult Honsha (Inst), Chugai Pharma (Inst), Takeda Pharmaceuticals (Inst), Eli Lilly (Inst), Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Daiichi Sankyo (Inst), Johnson & Johnson (Inst), Covidien (Inst), Otsuka (Inst), Nippon Kayaku (Inst), Tsumura (Inst), Asahi Kasei (Inst), Eisai (Inst), Kyowa Hakko Kirin (Inst), Astellas Pharma (Inst), Toyama Chemical (Inst), KCI Pharma (Inst), Abbott Laboratories (Inst)
Travel, Accommodations, Expenses: Taiho Pharmaceutical, Chugai Pharma, Yakult Honsha, Takeda Pharmaceuticals, Eli Lilly, Daiichi Sankyo, Ono Pharmaceutical, Merck Serono, EA Pharma, Johnson & Johnson, Covidien, Bayer Yakuhin, Eisai, Olympus, Terumo, Bristol-Myers Squibb Japan, Otsuka, Sanofi, Denka, Nippon Kayaku, Merck Sharp & Dohme
Takashi Kuwayama
No relationship to disclose
Seigo Nakamura
Honoraria: AstraZeneca, Pfizer, Chugai Pharma, Taiho Pharmaceutical
Chie Toshikawa
No relationship to disclose
Hideko Yamauchi
Honoraria: Taiho Pharmaceutical, AstraZeneca, Shionogi Pharma, Elsevier Japan, Chugai Pharma, Novartis, Pfizer, Aragan Japan, Towa Pharmaceutical
Consulting or Advisory Role: AstraZeneca
Teruo Yamauchi
Honoraria: AstraZeneca (I), Elsevier (I), Shionogi Pharma (I), Taiho Pharmaceutical (I), Chugai Pharma (I), Novartis (I), Pfizer (I)
Consulting or Advisory Role: AstraZeneca (I)
Koji Kaneko
No relationship to disclose
Chizuko Kanbayashi
No relationship to disclose
Nobuaki Sato
Honoraria: Chugai Pharmaceutical, AstraZeneca, Eisai, Pfizer, Taiho Pharmaceutical
Yasuo Miyoshi
Speakers’ Bureau: Chugai Pharma, AstraZeneca, Novartis, Pfizer, Taiho Pharmaceutical, Takeda Pharmaceuticals, Eli Lilly, Kyowa Hakko Kirin, Merck Sharp & Dohme, Daiichi Sankyo
Research Funding: Chugai Pharma (Inst), Novartis (Inst), Pfizer (Inst), Taiho Pharmaceutical (Inst), Takeda Pharmaceuticals (Inst), Eli Lilly (Inst), Merck Sharp & Dohme (Inst), Daiichi Sankyo (Inst)
Junko Tsuchida
No relationship to disclose
Masato Nakajima
No relationship to disclose
Yoshifumi Shimada
No relationship to disclose
Hiroshi Ichikawa
No relationship to disclose
Stephen Lyle
Employment: KEW
Stock and Other Ownership Interests: KEW
Consulting or Advisory Role: Pfizer
Kazuaki Takabe
No relationship to disclose
Shujiro Okuda
No relationship to disclose
Toshifumi Wakai
No relationship to disclose
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Sauvaget C, Nishino Y, Konno R, et al. Challenges in breast and cervical cancer control in Japan. Lancet Oncol. 2016;17:e305–e312. doi: 10.1016/S1470-2045(16)30121-8. [DOI] [PubMed] [Google Scholar]
- 3.Tsuchida J, Nagahashi M, Rashid OM, et al. At what age should screening mammography be recommended for Asian women? Cancer Med. 2015;4:1136–1144. doi: 10.1002/cam4.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irvin WJ, Jr, Carey LA. What is triple-negative breast cancer? Eur J Cancer. 2008;44:2799–2805. doi: 10.1016/j.ejca.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 5.Bianchini G, Balko JM, Mayer IA, et al. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao H, He G, Yan S, et al. Triple-negative breast cancer: Is there a treatment on the horizon? Oncotarget. 2017;8:1913–1924. doi: 10.18632/oncotarget.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [Erratum: Nature 491:288, 2012] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexandrov LB, Stratton MR. Mutational signatures: The patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev. 2014;24:52–60. doi: 10.1016/j.gde.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ju YS, Alexandrov LB, Gerstung M, et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. eLife. 2014;3:3. doi: 10.7554/eLife.02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shlien A, Raine K, Fuligni F, et al. Direct transcriptional consequences of somatic mutation in breast cancer. Cell Reports. 2016;16:2032–2046. doi: 10.1016/j.celrep.2016.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertucci F, Chaffanet M, Birnbaum D. An ICGC major achievement in breast cancer: A comprehensive catalog of mutations and mutational signatures. Linchuang Zhongliuxue Zazhi. 2017;6:4. doi: 10.21037/cco.2016.11.01. [DOI] [PubMed] [Google Scholar]
- 15.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowy DR, Collins FS. Aiming high: Changing the trajectory for cancer. N Engl J Med. 2016;374:1901–1904. doi: 10.1056/NEJMp1600894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bando H, Takebe N. Perspectives on research activity in the USA on cancer precision medicine. Jpn J Clin Oncol. 2016;46:106–110. doi: 10.1093/jjco/hyv162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chamberlin MD, Bernhardt EB, Miller TW. Clinical implementation of novel targeted therapeutics in advanced breast cancer. J Cell Biochem. 2016;117:2454–2463. doi: 10.1002/jcb.25590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagahashi M, Wakai T, Shimada Y, et al. Genomic landscape of colorectal cancer in Japan: Clinical implications of comprehensive genomic sequencing for precision medicine. Genome Med. 2016;8:136. doi: 10.1186/s13073-016-0387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan DS, Mok TS, Rebbeck TR. Cancer genomics: Diversity and disparity across ethnicity and geography. J Clin Oncol. 2016;34:91–101. doi: 10.1200/JCO.2015.62.0096. [DOI] [PubMed] [Google Scholar]
- 21.Dietze EC, Sistrunk C, Miranda-Carboni G, et al. Triple-negative breast cancer in African-American women: Disparities versus biology. Nat Rev Cancer. 2015;15:248–254. doi: 10.1038/nrc3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edge SB, Compton CC, Fritz AG, et al. AJCC Cancer Staging Manual. ed 7. New York, NY: Springer; 2010. [Google Scholar]
- 23.Barnes DM, Harris WH, Smith P, et al. Immunohistochemical determination of oestrogen receptor: Comparison of different methods of assessment of staining and correlation with clinical outcome of breast cancer patients. Br J Cancer. 1996;74:1445–1451. doi: 10.1038/bjc.1996.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 25.Eifert C, Pantazi A, Sun R, et al. Clinical application of a cancer genomic profiling assay to guide precision medicine decisions. Per Med. 2017;14:309–325. doi: 10.2217/pme-2017-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 27.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology: Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim TM, Laird PW, Park PJ. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell. 2013;155:858–868. doi: 10.1016/j.cell.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Kuraya K, Schraml P, Sheikh S, et al. Predominance of high-grade pathway in breast cancer development of Middle East women. Mod Pathol. 2005;18:891–897. doi: 10.1038/modpathol.3800408. [DOI] [PubMed] [Google Scholar]
- 31.Balko JM, Giltnane JM, Wang K, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014;4:232–245. doi: 10.1158/2159-8290.CD-13-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross JS, Ali SM, Wang K, et al. Comprehensive genomic profiling of inflammatory breast cancer cases reveals a high frequency of clinically relevant genomic alterations. Breast Cancer Res Treat. 2015;154:155–162. doi: 10.1007/s10549-015-3592-z. [DOI] [PubMed] [Google Scholar]
- 33.Parsons HA, Beaver JA, Cimino-Mathews A, et al. Individualized Molecular Analyses Guide Efforts (IMAGE): A prospective study of molecular profiling of tissue and blood in metastatic triple-negative breast cancer. Clin Cancer Res. 2017;23:379–386. doi: 10.1158/1078-0432.CCR-16-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagahashi M, Shimada Y, Ichikawa H, et al. Formalin-fixed paraffin-embedded sample conditions for deep next generation sequencing. J Surg Res. 2017;220:125–132. doi: 10.1016/j.jss.2017.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lips EH, Michaut M, Hoogstraat M, et al. Next generation sequencing of triple negative breast cancer to find predictors for chemotherapy response. Breast Cancer Res. 2015;17:134. doi: 10.1186/s13058-015-0642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nik-Zainal S, Davies H, Staaf J, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palma G, Frasci G, Chirico A, et al. Triple negative breast cancer: looking for the missing link between biology and treatments. Oncotarget. 2015;6:26560–26574. doi: 10.18632/oncotarget.5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuza K, Nagahashi M, Watanabe S, et al. Hypermutation and microsatellite instability in gastrointestinal cancers. Oncotarget. 2017;8:112103–112115. doi: 10.18632/oncotarget.22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peshkin BN, Alabek ML, Isaacs C. BRCA1/2 mutations and triple negative breast cancers. Breast Dis. 2010;32:25–33. doi: 10.3233/BD-2010-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner NC, Reis-Filho JS. Tackling the diversity of triple-negative breast cancer. Clin Cancer Res. 2013;19:6380–6388. doi: 10.1158/1078-0432.CCR-13-0915. [DOI] [PubMed] [Google Scholar]
- 42.Radovich M, Clare SE, Atale R, et al. Characterizing the heterogeneity of triple-negative breast cancers using microdissected normal ductal epithelium and RNA-sequencing. Breast Cancer Res Treat. 2014;143:57–68. doi: 10.1007/s10549-013-2780-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Millis SZ, Gatalica Z, Winkler J, et al. Predictive biomarker profiling of > 6000 breast cancer patients shows heterogeneity in TNBC, with treatment implications. Clin Breast Cancer. 2015;15:473–481.e3. doi: 10.1016/j.clbc.2015.04.008. [DOI] [PubMed] [Google Scholar]