Abstract
The genetic basis of antineutrophil cytoplasmic antibody, an important biomarker of inflammatory bowel disease (IBD), has never been thoroughly examined on a genome-wide scale. In this study, we performed a 2-stage genome-wide association study (GWAS) on antineutrophil cytoplasmic antibody in IBD cases. In the 2959 IBD cases in the discovery stage, we observed an association between a variant in the gene TNFRSF1B with antineutrophil cytoplasmic antibody level (rs5745994, minor allele frequency = 0.028, beta = 18.12, 95% CI, 11.82–24.22, P = 1.89 × 10−8). This association was replicated in an independent cohort of 419 IBD cases (beta = 16.91, 95% CI, 6.13–27.69, P = 2.38 × 102 ). With a Q-value of 0.036, we performed a fixed-effect meta-analysis for the association of rs5745994 in both cohorts and observed a stronger association signal (beta= 17.81, 95% CI, 12.36–23.25, P = 8.97 × 102 ). TNFRSF1B gene codes for tumor necrosis factor (TNF) receptor 2 (TNFR2), thereby we examined the reported TNFRSF1B variant with serum TNFR2 level. We observed a negative association with serum TNFR2 level being 8.23 EU/mL in carriers and 9.12 EU/mL in noncarriers (P = 0.033). This finding indicates the functional role of identified TNFRSF1B variant in IBD serology and may be reflective of the underlying biological mechanisms that determine clinical expression and/or response to certain therapies.
Keywords: GWAS, inflammatory bowel diseases, ANCA, TNFRSFIB
The inflammatory bowel diseases (IBDs) are a significant cause of morbidity in the United States with more than 1 million Americans affected by either ulcerative colitis (UC) or Crohn’s disease (CD),1–3 2 of the most common forms of IBD. It is hypothesized that IBD occurs due to an inappropriate immune response to normal enteric flora in genetically susceptible individuals4 and serological responses to microbial or autoantigens are seen at higher prevalence in IBD.5 A number of serological markers have been used in clinical practice, including atypical perinuclear antineutrophil cytoplasmic antibody (ANCA).6,7 Studies have investigated their potential value in the diagnosis of IBD8 and distinguishing CD from UC.9 Furthermore, a number of studies have demonstrated that ANCA is associated with complications related to IBD, including erythema nodosum10 and the development of chronic pouchitis.11 Importantly, a number of studies have demonstrated that ANCA is associated with nonresponse to anti–tumor necrosis factor (TNF) treatment although the mechanism of this association is unclear.12–14
Previous studies have suggested an association with ANCA in IBD and genetic variants within the HLA region,15–17 and the presence of ANCA is familial.15,17 Given the fact that ANCA is an important “intermediate” phenotype in IBD, understanding the genetic basis of ANCA can provide insight to the pathogenesis of a subset of IBD. Furthermore, with the potential clinical associations with ANCA as described above, genetic associations with ANCA may provide important insights to additional mechanisms, especially with respect to nonresponse to anti-TNF agents. Previous ANCA studies have been limited both by density of genotyping and by sample size.14–16 In this study, we performed a 2-stage genome-wide association study (GWAS) on ANCA in IBD cases. Genetic variation in TNFRSF1B region was associated with ANCA level in both discovery and replication cohorts.
METHODS
Subjects
Cedars-Sinai Cohort
Details of subject recruitment have been described previously.18–21 Briefly, 3110 patients with IBD were recruited at the Cedars-Sinai Inflammatory Bowel Disease Centers from 1985 to 2010. The diagnosis of each patient was based on standard endoscopic, histologic, and radiographic feature as previously described.22 Blood samples were collected at the time of enrollment. All study participants gave written informed content and the study protocol and data collection, as well as DNA preparation/genotyping and antibody measurement, was approved by the Cedars-Sinai Medical Center’s Institutional Review Board.
Mount Sinai Hospital Cohort
A total of 1853 IBD cases were recruited and similarly characterized at Mount Sinai Hospital in Toronto, Canada. All subjects provided written consent after institutional review board’s approval.23 The diagnosis of each patient was based on standard endoscopic, histologic, and radiographic feature.
ANCA Level Measurement
ANCA levels on serum from subjects from both centers were measured by enzyme-linked immunosorbent assay as previously described.22 All sera were analyzed in a blinded fashion at Cedars-Sinai Medical Center. Antibody levels were determined and results expressed as enzyme-linked immunosorbent assay units (EU/mL) as compared with a positive control.22 Qualitative positivity to ANCA was defined as being greater than cutoff values greater than 2 SDs above mean control titers.
Whole-Genome Genotyping
Cedars-Sinai Cohort
Genotyping was performed at Cedars-Sinai Medical Center using Illumina whole-genome arrays per manufacturer’s protocol (Illumina, San Diego, CA). The discovery cohort was genotyped on 3 platforms, including Illumina HumanCNV370-Quad (830 subjects), Human610-Quad (1037 subjects), and HumanOmniExpress (1243 subjects) arrays (total 3110 independent subjects). Average genotyping call rates for samples that passed quality control (QC) were 99.86% (HumanCNV370-Quad), 99.83% (Human610-Quad), and 99.85% (HumanOmniExpress). One to two percent of samples were genotyped in replicate and yielded average 99.99% concordance for genotypes called. Optimal allele-calling was verified by manual review of top associated single-nucleotide polymorphisms (SNPs).
A stringent QC procedure was applied to the GWAS data. Of the 3110 subjects genotyped, 10 were removed because of high missing rate (>2%), 27 were removed because of cryptic relatedness (Pi_Hat > 0.05), 3 were removed because sample either withdrew from the study or was later reclassified as non-IBD, leaving 3070 individuals for further analysis. Seventy-six individuals identified as nonwhites by principal component analysis were also removed. A total of 2959 of the 2994 genotyped subjects had ANCA status and were thereby included in the analyses.
Mount Sinai Hospital Cohort
A total of 1853 patients with IBD from the Mount Sinai Hospital Cohort in Toronto were genotyped on Illumina HumanOmniExpress array. Average genotyping call rates for samples that passed QC were 99.88%. Thirty-two samples genotyped in replicate and yielded average 99.99% concordance for genotypes called. After similar QC procedures as the discovery cohort, 1834 subjects remain in the cleaned data set, of which 419 had ANCA status available measured in the same laboratory as the discovery cohort. The 419 individuals with consistent ANCA measurements were included in this study as replication cohort.
Imputation
To consolidate data from different genotyping platforms, imputation was performed using a hidden Markov model-based algorithm available in the IMPUTE224 software package with Phase 2 HapMap genotypes as the Ref. 25. In the imputed data set, we retained only imputed SNPs with minor allele frequency (MAF) > 0.01 and quality score >0.80 in all 3 Illumina platforms, leaving approximately 2.01M SNPs for analyses.
Validation of Top-Hit SNP rs5745994
The most significant novel association observed in the genome-wide association analysis was with an imputed SNP (rs5745994). To validate the imputation, this SNP was included as custom content on the Illumina HumanExome+ array. BeadChip and genotyped as part of a larger project at Cedars-Sinai Medical Center following manufacturer’s protocol (Illumina, San Diego, CA). Genotype clusters for rs5745994 were visualized to ensure accurate allele-calling. The average genotyping rate of samples in the project that passed genotyping QC was 99.98%. Two hundred seventy-three samples performed in replicate as controls yielded 99.9963% concordance for genotypes called.
Measure of TNFRI and TNFR2 Levels
TNFR1 and TNFR2 concentrations were assessed in serum by commercially available enzyme-linked immunosorbent assay kits according to the manufacturer specified protocol (Biosource Invitrogen, KAC1761, KAC1771). The assay laboratory at Cedars-Sinai was blinded to subject status. Concentrations are reported in nanogram per milliliter.
Statistical Analysis
ANCA level in IBD cases is not normally distributed and the traditional method for analyzing skewed data, the nonparametric Mann–Whitney U test, is known to have lower efficiency and cannot incorporate covariates in analyses.26 Therefore, we used linear regression with the nontransformed ANCA level as outcome and for top SNPs, a permutation test was used to retain the type I error rate. The top 3 principal components and genotyping platform (in the discovery cohort) were included in the analysis as covariates to control for potential confounding. SNPTEST 2.2027 and R3.0228 were used in the statistical analysis. Similarly, permutation test was also performed in the analysis comparing circulating levels of TNFR I and TNFR II in carriers and noncarriers of the TNFRSF1B patients.
RESULTS
Demographic Characteristics
Table 1 shows the demographic characteristics of the 2959 individuals included in the final analyses of the discovery cohort of 1653 (55.86%) CD, 1193 (40.31%) UC, and 113 (3.82%) IBD-unclassified. The average age of disease onset was 25.65 ± 14.56 years old and 1513 (51.13%) are men. Median ANCA levels were 13.21 in CD, 36.00 in UC, and 18.93 in all IBD. A total of 433 CD (26.19%), 788 UC (66.05%), and 1278 all IBD (43.19%) were ANCA positive.
TABLE 1.
Demographic Characters of the Discovery Cohort
| CD | UC | All IBD | |
|---|---|---|---|
| N | 1653 | 1193 | 2959a |
| Sex (M/F) | 842/811 | 608/585 | 1513/1446 |
| Age of diagnosis | 23.64 ± 13.73 | 28.19 ± 15.05 | 25.65 ± 14.56 |
| ANCA level (median) | 13.21 | 36.00 | 18.93 |
| ANCA +/− | 433/1220 | 788/405 | 1278/1681 |
One hundred thirteen IBD undetermined patients were also included.
Association with ANCA Level
Overall genomic inflation factor for the genome-wide association in the discovery cohort was 1.023 (see Fig. 1, Supplemental Digital Content 1, http://links.lww.com/IBD/B253). Figure 1 shows the Manhattan plot for association with ANCA level in the GWAS. Before performing permutation testing, we observed 2 signals that reached standard and stringent criteria for genome-wide significance threshold of 5.0 × 10−8. The most significant SNP (rs11757159, MAF = 0.326) is located in HLA-DRB6, with the minor allele C (frequency 0.33) associated with lower ANCA level (beta =−7.09, 95% CI, −9.27 to −4.91), with a permutation test P value of 2.55 × 10−10. The second signal achieving genome-wide significance was located on 1p36.22 (rs5745994, MAF = 0.028) which is located in the intron of TNFRSF1B. Individuals carrying the minor allele (C allele) have higher ANCA level (beta = 18.12, 95% CI, 11.82–24.22), with a P-value of 1.89 × 10−8 after permutation testing (Table 2). Additional haplotype-based analyses and conditional analyses for this region suggest that there is only 1 signal at this locus, tagged by rs5745994 (data not shown).
FIGURE 1.
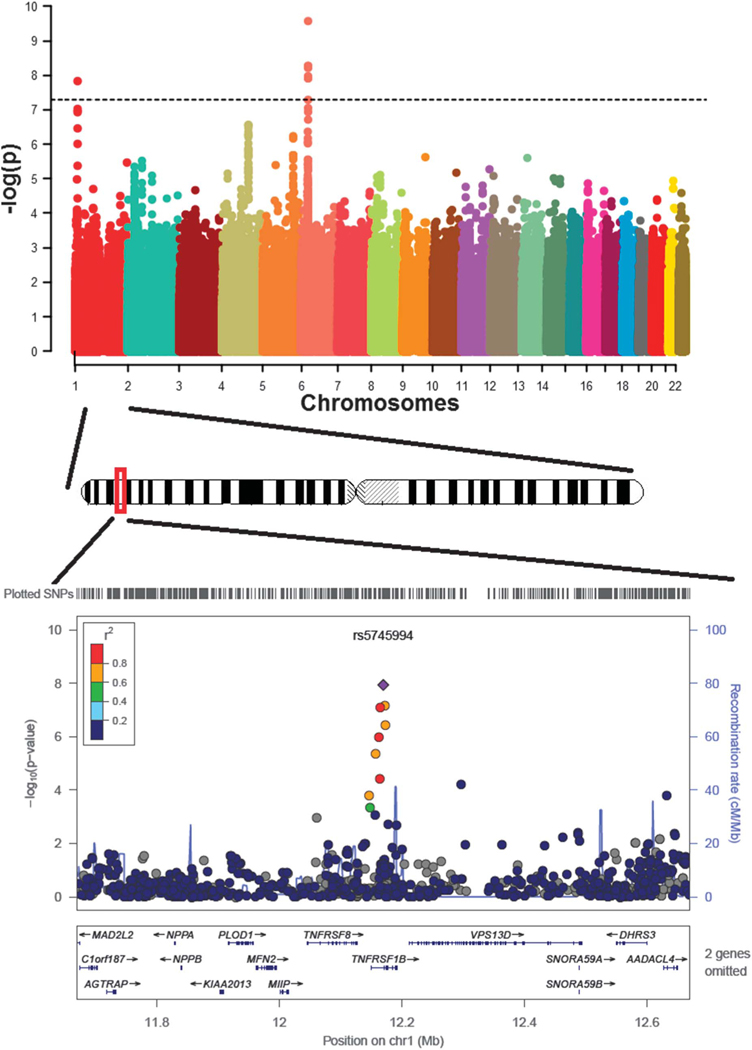
Genome-wide association with ANCA level in discovery cohort.
TABLE 2.
Top SNPs in GWAS Analysis
| Discovery Cohort (N = 2959) | Replication Cohort (N = 419) | Meta-Analysis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | CHR | Position | Gene | A1 | A2 | MAF | Beta (95% Cl) | Pa | MAF | Beta (95% Cl) | Pa | Beta (95% Cl) | Pa |
| rsl 1757159 | 6 | 32.628250 | HLA-DRB6 | T | C | 0.326 | −7.09 (−9.27 to −4.91) | 2.55 × 10−10 | 0.294 | −2.30 (−6.28 to 1.69) | 0.262 | −5.99 (−7.91 to −4.07) | 3.25 × 10−8 |
| rs5745994 | 1 | 12.169715 | TNFRSF1B | T | C | 0.028 | 18.12 (11.86 to 24.38) | 1.89 × 10−8 | 0.031 | 16.91 (6.13 to 27.69) | 2.38 × 10−3 | 17.81 (12.36 to 23.25) | 8.97 × 10−10 |
P values after permutation test.
CHR, chromosome; MAF, minor allele frequency.
We further examined the association of the novel signal in TNFRSF1B separately in CD and UC. The signal is observed in both CD and UC, with a stronger effect observed in UC (CD: beta = 11.14, 95% CI, 4.67–17.62, P = 1.24 × 10−3; UC: beta = 23.83, 95% CI, 12.88–34.78, P = 8.42 × 10−5).
Validation of the TNFRSF1B SNP in ExomeChip
As the TNFRSF1B SNP (rs5745994) was an imputed SNP, we validated the imputation by incorporating this SNP as an SNP in the customized content in our ExomeChip genotyping efforts. Concordance rate between imputed and genotyped data for 2841 individuals with both GWAS and ExomeChip data was 99.64%. As expected, association analysis using only genotyped SNPs yielded similar results (beta = 16.78, 95% CI, 10.16–23.40, P = 7.15 × 10−7).
Replication of the Top Signals
Using an additional cohort of 419 patients with IBD from Mount Sinai Hospital in Toronto in which ANCA levels were analyzed at Cedars-Sinai, we further examined the association of the TNFRSF1B with ANCA level (Table 2). We were able to confirm our original observation of an association between the C allele of rs5745994 and ANCA level, with similar effect magnitude (beta = 16.91, 95% CI, 6.13–27.69, P = 2.38 × 10−3). With a Q-value of 0.036, we performed a fixed-effect meta-analysis for the association of rs5745994 in both cohorts and observed a stronger association signal (beta = 17.81, 95% CI, 12.36–23.25, P = 8.97 × 10−10). The HLA signal observed in the discovery cohort failed to replicate (beta =−2.30, P = 0.262).
Association of the TNFRSF1B SNP with TNFR2 Level
Next, we tested for association between TNFRSF1B (rs5745994) and serum TNFR1 (as control) and TNFR2 levels in a subset of 239 patients with IBD, of which 58 carried the C allele of rs5745994 with the remainder being homozygous for the T allele. Figure 2 shows the association of rs5745994 with TNFR1 and TNFR2. TNFRSF1B was not associated with TNFR1 (median 2.99 EU/mL in C allele carriers compared with 3.06 EU/mL in noncarriers, P = 0.71). In contrast, the median TNFR2 was 8.23 EU/mL in carriers and 9.12 EU/mL in noncarriers (P = 0.033), indicating lower serum TNFR2 level in carriers of the C allele.
FIGURE 2.
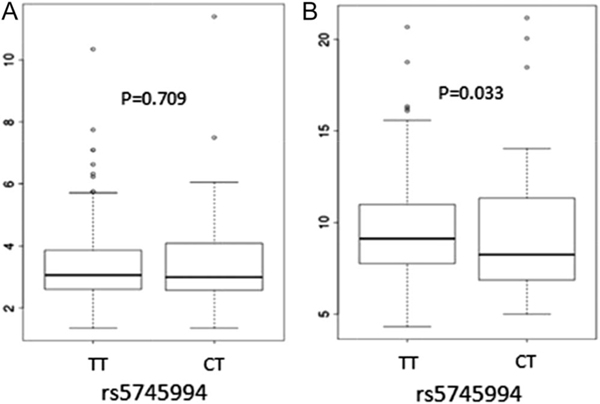
Association of the rs5745994 with serum TNFR I and TNFR II levels. A, Association of rs5745994 with TNFR I levels. B, Association of rs5745994 with TNFR II levels.
DISCUSSION
A number of studies have highlighted the potential association of ANCA with clinically relevant subsets of IBD and documenting higher ANCA levels in patients with IBD compared with non-IBD controls.6–8,11,13 Furthermore, ANCA is 1 of the few replicated parameters associated with likelihood of response of patients with IBD to anti-TNF treatment.12,14 Understanding the genetic basis of ANCA expression may help elucidate the underlying mechanisms of these associations. However, to date, genetic associations with ANCA expression in IBD have not been systematically evaluated using unbiased approaches in large-scale studies. In this study, we performed the first GWAS analysis of ANCA in patients with IBD. We identified and replicated a novel association of ANCA with TNFRSF1B which encodes the protein TNFR2. In addition, we demonstrated that the same variant is associated with lower serum levels of TNFR2.
TNFR2 is a membrane-bound receptor of TNF-α, a critical signaling protein in the immune system.29,30 In comparison with another TNF-α receptor TNFR1, which is expressed in almost all the cell types in human body,31 the expression of TNFR2 is largely restricted to immune-related cells,31,32 such as lymphocytes (CD4 and CD8 cells), endothelia cells, and thymocytes. TNF-α binding to TNFR2 triggers the recruitment of the adapter proteins TNF receptor-associated factor1 (TRAF1) and TNF receptor-associated factor2 (TRAF2),33,34 precipitating activation of the downstream cascade of nuclear factor-kappaB33,35 and c-Jun N-terminal kinase (JNK).35,36 This cascade results in proliferation of multiple immune-related cells, including cytotoxic T cells,31 thymocytes,37 mononuclear cells38 as well as Treg cells,39 indicating the complex and crucial role of TNFR2 in human immune response. TNFR2 is also reported to mediate slan dendritic cells (slanDCs) enhancement of NK-cell function40; promote suppressive activities of myeloid-derived suppressor cells41,42; and play a crucial auxiliary role to TNFR1 in sensitizing macrophages for the activation of the p38 mitogen-activated protein kinases (MAPK) and classical nuclear factor-kappaB proinflammatory signaling pathways.43
In this study, we identified a novel association between ANCA levels, TNFRSF1B genotype and decreased circulating TNFR2 levels, which may be reflective of the underlying biological mechanisms that determine clinical expression and/or response to certain therapies. Multiple reports have been published demonstrating that the presence of ANCA and/or levels of ANCA in the serum are associated with lack of response to anti-TNF therapy in patients with UC and CD.12,14,44,46 In CD, serum ANCA is related to an “ulcerative colitis-like” phenotype, characterized by left-sided colonic disease.47 In UC, high levels of ANCA are associated with more aggressive disease48,49 and onset of chronic pouchitis after ileal pull through anal anastomosis.11,49
Previous studies evaluating TNFRSF1B genotypes have shown a relationship with both CD and UC, but not with ANCA expression or response to anti-TNF therapeutics.50 The published studies on TNFRSF1B variants have shown an association only with higher levels of soluble TNFR2 in serum of familial rheumatoid arthritis and systemic lupus erythematosus.51–53 These associations were among the findings that signified the potential therapeutic benefit of etanercept (soluble P75, TNFR2) to treat rheumatoid arthritis and psoriasis by binding soluble TNF. Etanercept is ineffective in CD and also does not induce apoptosis in vivo or in vitro mucosal experiments suggesting that apoptosis induction is important for biological and clinical responses to antiTNF therapeutics in CD and UC.
Evidence that ANCA is generated by mucosal B cells suggests that serum ANCA is a result of reactivity to local antigens.54 We have shown that the antigen for IBD-associated ANCA is nuclear and reactivity can be eliminated by pretreating neutrophils with DNase (ANCA),55 which suggests that ANCA could be a marker for increased nuclear destruction within the mucosa involving release of nuclear antigens potentially related to ongoing apoptotic activity.56 The possibility that ANCA is related to apoptosis is especially interesting because 1 of the main mechanisms by which anti-TNF inhibits mucosal inflammation is induction of T-cell apoptosis.56,57 A study parsed out the details of this mechanism.56 The data demonstrated that the ultimate level of apoptosis is dependent on a balance of pro versus antiapoptotic signal generating molecules that induce or protect against apoptosis. Membrane TNF, present on macrophages, preferentially binds to TNFR2 as opposed to TNFR1 on activated CD4 T cells.56 Downstream signaling of TNFR2 activates NFKB, which induces intracellular molecules and soluble cytokines that protect the cell from proapoptotic signals and raise the threshold for apoptosis in CD4 T cells. The result is increasing numbers of activated mucosal T cells in patients with IBD.58 Therapeutic antibodies to TNF bind transmembrane TNF, potentially preventing TNFR2 signaling that lowers the apoptotic threshold and makes these cells more susceptible to ongoing apoptosis, resulting in decreased numbers of mucosal effector T cells.56 Thus, high ANCA levels and lower levels of TNFR2 (reflected by lower circulating TNFR2 levels) suggest ongoing mucosal apoptosis. Therefore, treating this population with anti-TNF will not further enhance apoptosis and a therapeutic that targets different mechanisms would be indicated.
Our study has some limitations. First, the discovery cohort consists of 3 different genotyping platforms (Illumina 370K, 610K and Omni Express). Although imputation was performed in the 3 cohorts and stringent QC procedures were applied, we needed to include the genotyping platform as a covariate to control for potential confounding. Thus, the top SNPs were verified through direct genotyping on the ExomeChip which guarantees the genotyping and imputation quality of our major finding. Moreover as mentioned above, the sample size of the replication cohort is small, which might be the reason why the HLA SNP observed in the discovery cohort failed to replicate.
In summary, we observed an association between a variant in TNFRSF1B gene and ANCA levels in patients with IBD. This association is replicated in an independent cohort. TNFRSF1B codes for TNFR2 which plays an important role in immune response. In functional analysis, we observed an association between the TNFRSF1B variant and decreased circulating TNFR2 level, which indicates the functional role of the identified variant and moreover, might reflect the underlying biological mechanisms that might have important implication in clinical practice.
Supplementary Material
Acknowledgments
Supported by F. Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute and NIH NIDDK P01DK046763.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.ibdjournal.org).
The authors have no conflict of interest to disclose.
REFERENCES
- 1.Loftus EV Jr. Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology. 2004; 126:1504–1517. [DOI] [PubMed] [Google Scholar]
- 2.Loftus EV Jr. The burden of inflammatory bowel disease in the United States: a moving target? Clin Gastroenterol Hepatol. 2007;5:1383–1384. [DOI] [PubMed] [Google Scholar]
- 3.Kappelman MD, Rifas-Shiman SL, Kleinman K, et al. The prevalence andgeographic distribution of Crohn’s disease and ulcerative colitis in the United States. Clin Gastroenterol Hepatol. 2007;5:1424–1429. [DOI] [PubMed] [Google Scholar]
- 4.Bamias G, Cominelli F. Imunopathogenesis of inflammatory bowel disease: current concepts. Curr Opin Gastroenterol. 2007;23:365–369. [DOI] [PubMed] [Google Scholar]
- 5.Papp M, Norman GL, Altorjay I, et al. Utility of serological markers ininflammatory bowel diseases: gadget or magic? World J Gastroenterol. 2007;13:2028–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montanelli A, Mainardi E, Vagni A, et al. Immunological markers anti-saccharomyces cerevisiae antibodies (ASCA) and anti-neutrophil cytoplasmic antibodies (ANCA) in inflammatory bowel disease: a helpful diagnostic tool. Minerva Gastroenterol Dietol. 2005;51:201–207. [PubMed] [Google Scholar]
- 7.Lewis JD. The utility of biomarkers in the diagnosis and therapy of inflammatory bowel disease. Gastroenterology. 2011;140:1817–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reese GE, Constantinides VA, Simillis C, et al. Diagnostic precision of anti-saccharomyces cerevisiae antibodies and perinuclear antineutrophil cytoplasmic antibodies in inflammatory bowel disease. Am J Gastroenterol. 2006;101:2410–2422. [DOI] [PubMed] [Google Scholar]
- 9.Joossens S, Reinisch W, Vermeire S, et al. The value of serologic markersin indeterminate colitis: a prospective follow-up study. Gastroenterology. 2002;122:1242–1247. [DOI] [PubMed] [Google Scholar]
- 10.Turkcapar N, Toruner M, Soykan I, et al. The prevalence of extraintestinal manifestations and HLA association in patients with inflammatory bowel disease. Rheumatol Int. 2006;26:663–668. [DOI] [PubMed] [Google Scholar]
- 11.Fleshner P, Ippoliti A, Dubinsky M, et al. Both preoperative perinuclear antineutrophil cytoplasmic antibody and anti-CBir1 expression in ulcerative colitis patients influence pouchitis development after ileal pouch-anal anastomosis. Clin Gastmentenol Hepatol. 2008;6:561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor KD, Plevy SE, Yang H, et al. ANCA pattern and LTA haplotyperelationship to clinical responses to anti-TNF antibody treatment in Crohn’s disease. Gastroenterology. 2001;120:1347–1355. [DOI] [PubMed] [Google Scholar]
- 13.Esters N, Vermeire S, Joossens S, et al. Serological markers for predictionof response to anti-tumor necrosis factor treatment in Crohn’s disease. Am J Gastroenterol. 2002;97:1458–1462. [DOI] [PubMed] [Google Scholar]
- 14.Yang H, Rotter JI, Toyoda H, et al. Ulcerative colitis: a genetically heterogeneous disorder defined by genetic and subclinical markers. J Clin Invest. 1993;92:1080–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YT, Sung JJ, Poon P, et al. Association of HLA class-II genes andanti-neutrophil cytoplasmic antibodies in Chinese patients with inflammatory bowel disease. Scand J Gastroenterol. 1998;33:623–627. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto-Furusho JK, Uscanga-Domínguez L, Lopez-Martinez A, et al. Association of the HLA-DRB1*0701 allele with perinuclear antineutrophil cytoplasmatic antibodies in Mexican patients with severe ulcerative colitis. World J Gastroenterol. 2006;12:1617–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shanahan F, Duerr RH, Rotter JI, et al. Neutrophil autoantibodies inulcerative colitis: familial aggregation and genetic heterogeneity. Gastroenterology. 1992;103:456–461. [DOI] [PubMed] [Google Scholar]
- 18.Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysisincreases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGovern DP, Jones MR, Taylor KD, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Hum Mol Genet. 2010;19:3468–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haritunians T, Taylor KD, Targan SR, et al. Genetic predictors of medically refractive ulcerative colitis, Inflamm Bow Dis. 2010;16:1830–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mow WS, Vasiliauskas EA, Lin YC, et al. Association of antibody responses to microbial antigens and complications of small bowel Crohn’s disease. Gastroenterology. 2004;126:414–424. [DOI] [PubMed] [Google Scholar]
- 23.Muise AM, Xu W, Guo CH, et al. NADPH oxidase complex and IBDcandidate gene studies: identification of a rare variant in NCF2 that results in reduced binding to RAC2. Gut. 2012;61:1028–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchini J, Howie B. Genotype imputation for genome-wide associationstudies. Nat Rev Genet. 2010;11:499–511. [DOI] [PubMed] [Google Scholar]
- 25.International HapMap 3 Consortium. Integrating common and rare geneticvariation in diverse human populations. Nature. 2010;467:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehamnn EL. Elements of Large Sample Theory. New York: Springer; 1999:176. [Google Scholar]
- 27.Marchini J, Howie B, Myers S, et al. A new multipoint method forgenome-wide association studies via imputation of genotypes. Nat Genet. 2007;39:906–913. [DOI] [PubMed] [Google Scholar]
- 28.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. Available at: http://www.R-project.org/. Accessed July 16, 2014.
- 29.Brown GR, Thiele DL. Enhancement of MHC class I-stimulated alloresponses by TNF/TNF receptor (TNFR)1 interactions and of MHC class IIstimulated alloresponses by TNF/TNFR2 interactions. Eur J Immunol. 2000;30:2900–2907. [DOI] [PubMed] [Google Scholar]
- 30.Hamano R, Huang J, Yoshimura T, et al. TNF optimally activativesregulatory T cells by inducing TNF receptor superfamily members TNFR2, 4–1BB and OX40. Eur J Immunol. 2011;41:2010–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sedger LM, McDermott MF. TNF and TNF-receptors: from mediators ofcell death and inflammation to therapeutic giants—past, present and future. Cytokine Growth Factor Rev. 2014;25:453–472. [DOI] [PubMed] [Google Scholar]
- 32.Hu X, Li B, Li X, et al. Transmembrane TNF-α promotes suppressive activities of myeloid-derived suppressor cells via TNFR2. J Immunol. 2014;192:1320–1331. [DOI] [PubMed] [Google Scholar]
- 33.Blüml S, Scheinecker C, Smolen JS, et al. Targeting TNF receptors inrheumatoid arthritis. Int Immunol. 2012;24:275–281. [DOI] [PubMed] [Google Scholar]
- 34.Cabal-Hierro L, Rodríguez M, Artime N, et al. TRAF-mediated modulation of NF-kB AND JNK activation by TNFR2. Cell Signal. 2014;26:2658–2666. [DOI] [PubMed] [Google Scholar]
- 35.Yang S, Wang Y, Mei K, et al. TNFR2-IL-17RD heteromerization revealsa novel mechanism for NF-kB activation. J Biol Chem. 2015;290:861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W, Li H, Bocking AD, et al. Tumor necrosis factor stimulates matrixmetalloproteinase 9 secretion from cultured human chorionic trophoblast cells through TNF receptor 1 signaling to IKBKB-NFKB and MAPK1/3 pathway. Biol Reprod. 2010;83:481–487. [DOI] [PubMed] [Google Scholar]
- 37.Grell M, Becke FM, Wajant H, et al. TNF receptor type 2 mediatesthymocyte proliferation independently of TNF receptor type 1. Eur J Immunol. 1998;28:257–263. [DOI] [PubMed] [Google Scholar]
- 38.Nonaka K, Saio M, Suwa T, et al. Skewing the Th cell phenotype towardTh1 alters the maturation of tumor-infiltrating mononuclear phagocytes. J Leukoc Biol. 2008;84:679–688. [DOI] [PubMed] [Google Scholar]
- 39.Joedicke JJ, Myers L, Carmody AB, et al. Activated CD8+ T cells induceexpansion of Vb5+ regulatory T cells via TNFR2 signaling. J Immunol. 2014;193:2952–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tufa DM, Chatterjee D, Low HZ, et al. TNFR2 and IL-12 coactivationenables slanDCs to support NK-cell function via membrane-bound TNFa. Eur J Immunol. 2014;44:3717–3728. [DOI] [PubMed] [Google Scholar]
- 41.Polz J, Remke A, Weber S, et al. Myeloid suppressor cells require membrane TNFR2 expression for suppressive activity. Immun Inflamm Dis. 2014;2:121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mei S, Liu Y, Wu X, et al. TNF-a-mediated microRNA-136 induces differentiation of myeloid cells by targeting NFIA. J Leukoc Biol. 2016; 99:301–311. [DOI] [PubMed] [Google Scholar]
- 43.RuspG, SchmidM, McCanF, et al. TNFR2 increases the sensitivityof ligand-induced activation of the p38 MAPK and NF-kB pathways and signals TRAF2 protein degradation in macrophages. Cell Signal. 2014;26: 683–690. [DOI] [PubMed] [Google Scholar]
- 44.Ferrante M, Vermeire S, Katsanos KH, et al. Predictors of early responseto infliximab in patients with ulcerative colitis. Inflamm Bowel Dis. 2007; 13:123–128. [DOI] [PubMed] [Google Scholar]
- 45.Jurgens M, Laubender RP, Hartl F, et al. Disease activity, ANCA, andIL23R genotype status determine early response to infliximab in patients with ulcerative colitis. Am J Gastroenterol. 2010;105:1811–1819. [DOI] [PubMed] [Google Scholar]
- 46.Vasiliauskas EA, Plevy SE, Landers CJ, et al. Perinuclear antineutrophil cytoplasmic antibodies in patients with Crohn’s disease define a clinical subgroup. Gastroenterology. 1996;110:1810–1819. [DOI] [PubMed] [Google Scholar]
- 47.Sandborn WJ, Landers CJ, Tremaine WJ, et al. Antineutrophil cytoplasmic antibody correlates with chronic pouchitis after ileal pouch-anal anastomosis. Am J Gastroenterol. 1995;90:740–747. [PubMed] [Google Scholar]
- 48.Sandborn WJ, Landers CJ, Tremaine WJ, et al. Association of antineutrophil cytoplasmic antibodies with resistance to treatment of leftsided ulcerative colitis: results of a pilot study. Mayo Clinic Proc. 1996;71:431–436. [DOI] [PubMed] [Google Scholar]
- 49.Fleshner PR, Vasiliauskas EA, Kam LY, et al. High level perinuclear antineutrophil cytoplasmic antibody (ANCA) in ulcerative colitis patients before colectomy predicts the development of chronic pouchitis after ileal pouch-anal anastomosis. Gut. 2001;49:671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sashio H, Tamura K, Ito R, et al. Polymorphisms of the TNF gene and theTNF receptor superfamily member 1B gene are associated with susceptibility to ulcerative colitis and Crohn’s disease, respectively. Immunogenetics. 2002;53:1020–1027. [DOI] [PubMed] [Google Scholar]
- 51.Till A, Rosenstiel P, Krippner-Heidenreich A, et al. The Met-196 -> Arg variation of human tumor necrosis factor receptor 2 (TNFR2) affects TNF-alpha-induced apoptosis by impaired NF-kappaB signaling and target gene expression. J Bio Chem. 2005;280:5994–6004. [DOI] [PubMed] [Google Scholar]
- 52.Pierik M, Vermeire S, Steen KV, et al. Tumour necrosis factor-alphareceptor 1 and 2 polymorphisms in inflammatory bowel disease and their association with response to infliximab. Aliment Pharmacol Ther. 2004; 20:303–310. [DOI] [PubMed] [Google Scholar]
- 53.Canete JD, Albaladejo C, Hernandez MV, et al. Clinical significance of high levels of soluble tumour necrosis factor-alpha receptor-2 produced by alternative splicing in rheumatoid arthritis: a longitudinal prospective cohort study. Rheumatology. 2011;50:721–728. [DOI] [PubMed] [Google Scholar]
- 54.Targan SR, Landers CJ, Cobb L, et al. Perinuclear anti-neutrophil cytoplasmic antibodies are spontaneously produced by mucosal B cells of ulcerative colitis patients. J Immunol. 1995;155:3262–3267. [PubMed] [Google Scholar]
- 55.Vidrich A, Lee J, James E, et al. Segregation of ANCA antigenic recognition by DNase treatment of neutrophils: ulcerative colitis, type 1 autoimmune hepatitis, and primary sclerosing cholangitis. J Clin Immunol. 1995;15:293–299. [DOI] [PubMed] [Google Scholar]
- 56.Atreya R, Zimmer M, Bartsch B, et al. Antibodies against tumor necrosisfactor (TNF) induce T-cell apoptosis in patients with inflammatory bowel diseases via TNF receptor 2 and intestinal CD14(+) macrophages. Gastroenterology. 2011;141:2026–2038. [DOI] [PubMed] [Google Scholar]
- 57.Waetzig GH, Rosenstiel P, Arlt A, et al. Soluble tumor necrosis factor(TNF) receptor-1 induces apoptosis via reverse TNF signaling and autocrine transforming growth factor-beta1. FASEB J. 2005;19:91–93. [DOI] [PubMed] [Google Scholar]
- 58.Levine AD, Fiocchi C. Regulation of life and death in lamina propria Tcells. Semin Immunol. 2001;13:195–199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


