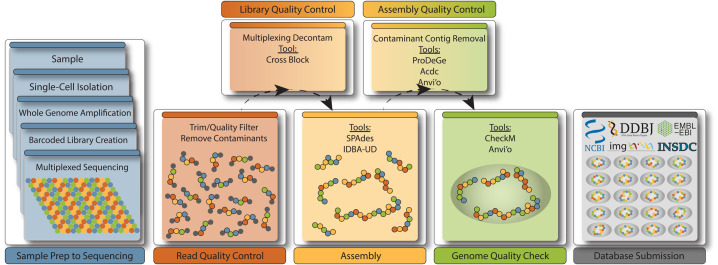
Figure 1. A schematic representation of the single-cell workflow with a focus on the analysis following sequencing.
Left panels with blue background represent the production of single-cell genomes, while the rest of the workflow relates specifically to the analysis of single-cell sequence data going from raw reads to public database submission. The bottom row of analysis boxes refers to the steps that are considered mandatory to any single-cell analysis pipeline, while the top row can be considered context-dependent. For example, if multiplexing was not performed, poolmate decontamination is not necessary (Library Quality Control). However, in nearly all cases, an SAG will benefit from contamination screening (Assembly Quality Control), as even the cleanest SAGs may contain a few contaminating contigs, and if not, this step can serve as validation of a clean SAG that is nearly ready for submission to the public databases.