Figure 4.
Comparison of Protein Phosphorylation and Kinase Activity in HGSC and Normal FT
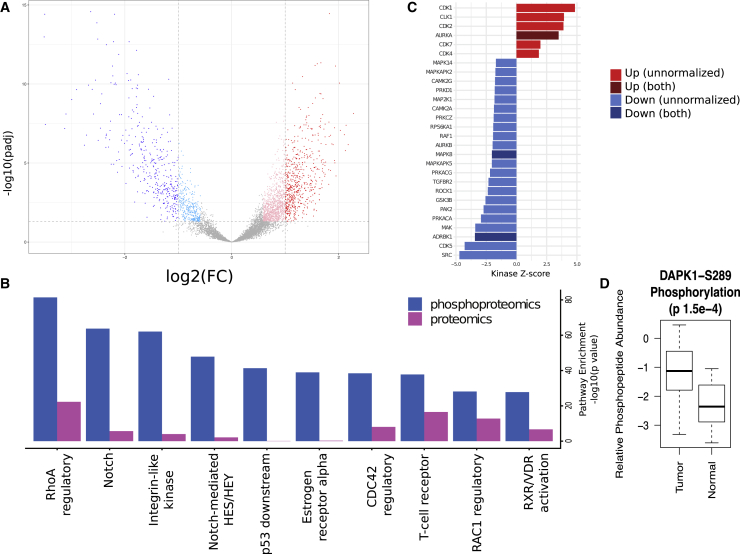
(A) Volcano plot of tumor-normal differences in peptide phosphorylation. Phosphopeptides with significantly increased or decreased (±2-fold change, Benjamini-Hochberg adjusted p < 0.05) abundance in tumors compared to normal FT are colored red and blue, respectively. The lighter colors are the significantly differential phosphopeptides at a lower fold-change threshold of ±1.5. An interactive version of this plot is available at https://doi.org/10.25584/cptac/1601822.
(B) Pathway components were compared (2-sided t test) between tumor and normal FT samples for differences in protein and phosphorylation abundance. The top 10 most significant pathways in terms of Benjamini-Hochberg adjusted p value for proteomics and phosphoproteomics data are shown. These pathways exhibit increased phosphorylation in tumors. The notch, integrin-like kinase, and RhoA regulatory pathways emerged as the most activated in this analysis.
(C) Inference of kinase activity in tumors compared to normal using KSEA on phosphopeptides for known kinase substrates. The bar plot shows the relative activity of kinases in tumors versus normal FT. The Z scores are a measure of the enrichment for phosphorylated substrates of a given kinase, with higher scores corresponding to greater activity in the tumors. Kinases with significant (FDR < 0.05) increases in tumors are in red, whereas those with significantly lower activity in tumors are in blue. Darker colors are used for kinases for which phosphorylation changes are independent of abundance changes.
(D) Comparison of DAPK1 phosphorylation at residue S289 in tumor and normal samples.
See also Figures S1 and S5.