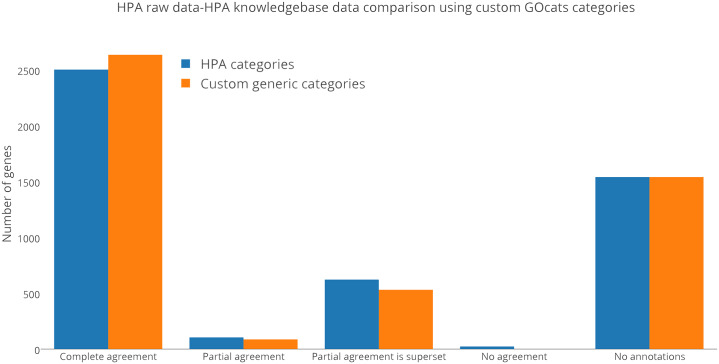
Fig 6. Comparison of HPA knowledgebase derived annotations to HPA experimental data.
Number of genes in the given agreement type when comparing gene product annotations assigned by HPA in the EMBL-EBI knowledgebase to those in The Human Protein Atlas’ raw experimental data. “Complete agreement” refers to genes where all subcellular locations derived from the knowledgebase and the HPA dataset matched, “partial agreement” refers to genes with at least one matching subcellular location, “partial agreement is superset” refers to genes where knowledgebase subcellular locations are a superset of the HPA dataset (these are mutually exclusive to the “partial agreement” category), "no agreement" refers to genes with no subcellular locations in common, and “no annotations” refers to genes in the experimental dataset that were not found in the knowledgebase. The more-generic categories used in panel B can be found in Table 3.