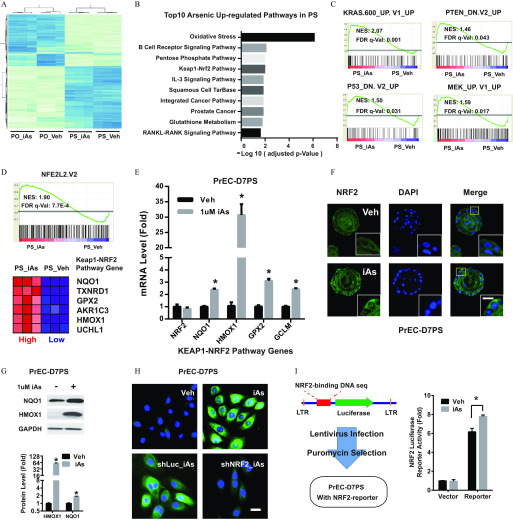
Figure 2.
Effects of arsenic on the NRF2 pathway in prostate stem-progenitor cells. (A) Heatmap showing the top 1,000 differentially expressed genes across four groups: prostate spheres arsenic (PS_Veh, PS_iAs), prostate organoids arsenic (PO-Veh, PO-iAs). Prostate spheres and organoids were derived from primary human prostate epithelial cells (PrEC), and cultured arsenic for 2 wk. (B) Top 10 iAs up-regulated pathways in prostate spheres, iAs up-regulated genes (, adjusted ) were sorted for pathway analysis with Wikipathway. (C) Enriched oncogenic pathways in PS_iAs group vs. PS_Veh group. Raw microarray data were subject to GSEA analysis using gene sets of oncogenic signatures. (D) GSEA data showing NRF2 pathway enrichment between PS-iAs and PS-Veh groups (top) and most regulated genes of NRF2 pathway (bottom). (E) mRNA level of NRF2 pathway genes in PS_Veh and PS_iAs group. Data shown are (); * vs. vehicle. (F) Representative NRF2 immunostaining showing nuclear translocation in PS upon iAs treatment for 7 d (PrEC-D7PS). . (G) Immunoblot of HMOX1 in PrEC-D7PS iAs treatment. Data shown are (); * vs. vehicle. (H) Representative HMOX1 immunostaining in day-7 spheres derived from primary human prostate epithelial cells (PrEC-D7PS) with indicated treatment, . iAs, arsenic; shLuc, negative control shRNA targeting luciferase gene; shNRF2, shRNA targeting NRF2 gene; Veh, vehicle control. . (I) NRF2-luciferase reporter assay showing luciferase activity in PrEC-D7PS iAs. Data shown are (); * vs. vehicle.