Extended Data Fig. 7. 3D super-resolution reconstructions of immunofluorescence-labeled Nup98 in COS-7 cells using INSPR and the in vitro method in biplane setup.
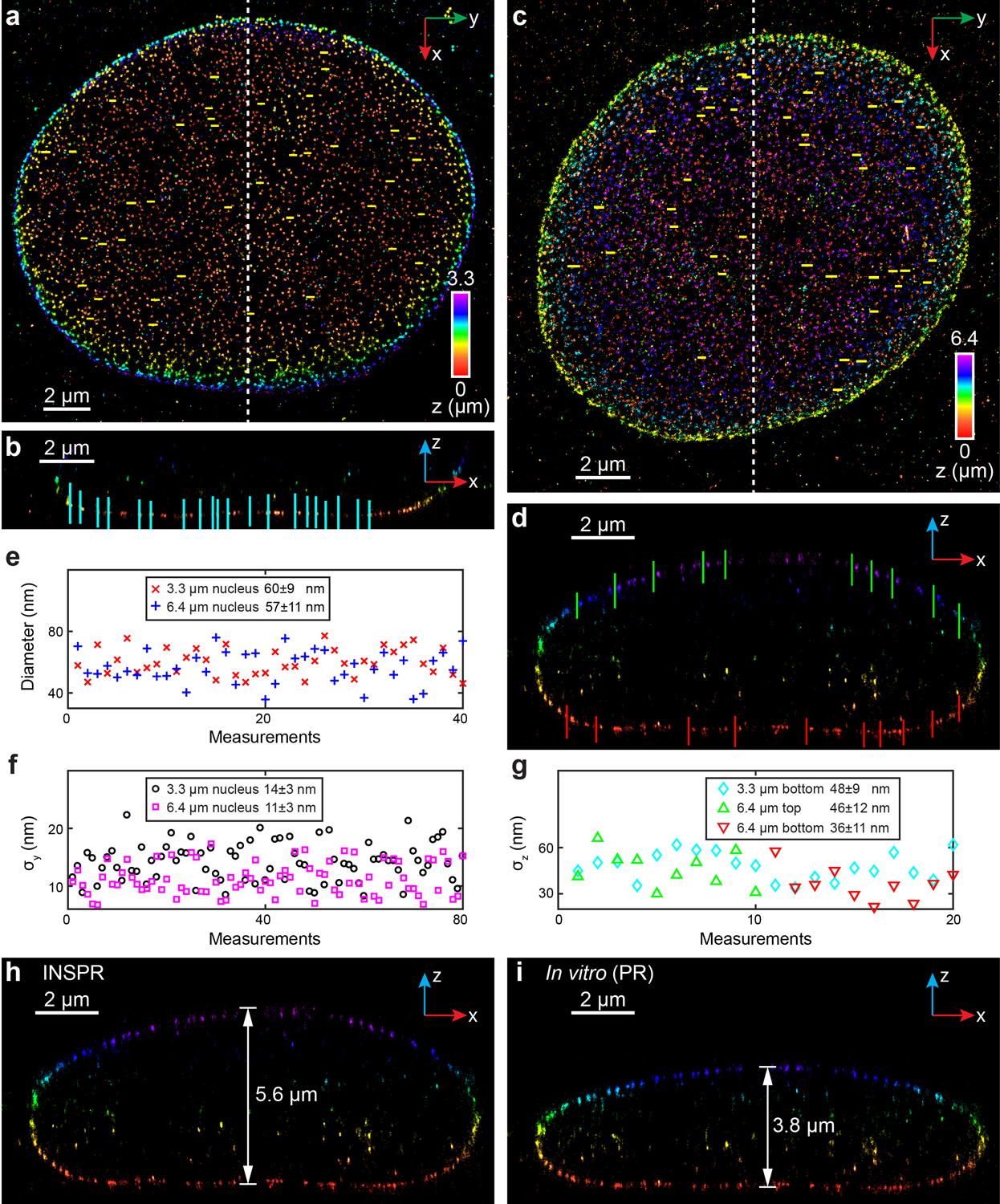
(a) x-y overview of the 3.3-μm-thick volume of the nucleus showing the positions of 40 typical Nup98 structures (yellow lines). (b) x-z slice along the white dashed line in (a), showing the positions of 20 typical Nup98 structures (cyan lines). (c) x-y overview of the 6.4-μm-thick entire nuclear envelope showing the positions of 40 typical Nup98 structures (yellow lines). (d) x-z slice along the white dashed line in (c), showing the positions of 10 typical Nup98 structures on the top (green lines) and bottom (red lines) surfaces. (e) Distribution of diameters measured from Nup98 structures in the x-y plane shown in (a,c). The diameter is 60±9 nm for the 3.3-μm-thick volume (mean±std, 40 measurements, red crosses), and 57±11 nm for the 6.4-μm-thick volume (40 measurements, blue plus signs). (f) Distribution of σy measured from Nup98 structures in the x-y plane shown in (a,c). σy is 14±3 nm for the 3.3-μm-thick volume (80 measurements, black circles), and 11±3 nm for the 6.4-μm-thick volume (80 measurements, magenta squares). (g) Distribution of σz measured from Nup98 structures in the x-z plane shown in (b,d). For the 3.3-μm-thick volume, σz is 48±9 nm (20 measurements, cyan diamonds). For the 6.4-μm-thick volume, σz is 46±12 nm for the top surface (10 measurements, green upward-pointing triangles), and 36±11 nm for the bottom surface (10 measurements, red downward-pointing triangles). (h,i) x-z slice along the white dashed line in (c), reconstructed using INSPR (h) and in vitro phase retrieval based on beads on the coverslip (in vitro (PR), i). The integration width of the x-z slices in (b,d,h,i) in the y direction is 500 nm.
