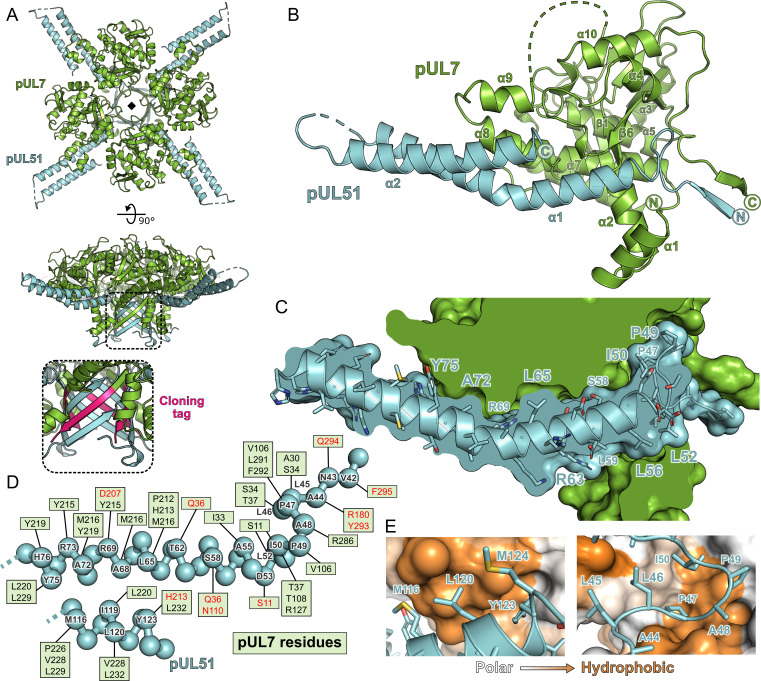
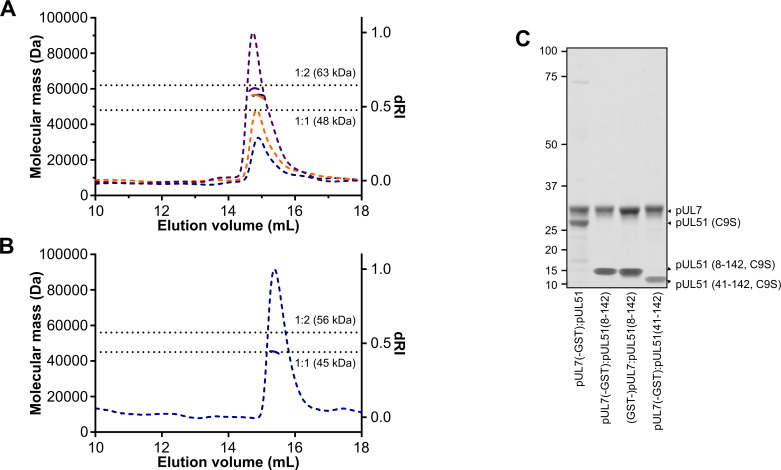
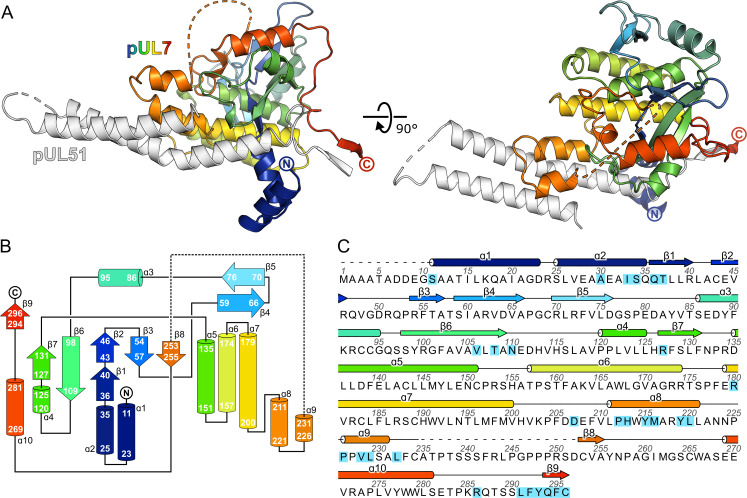
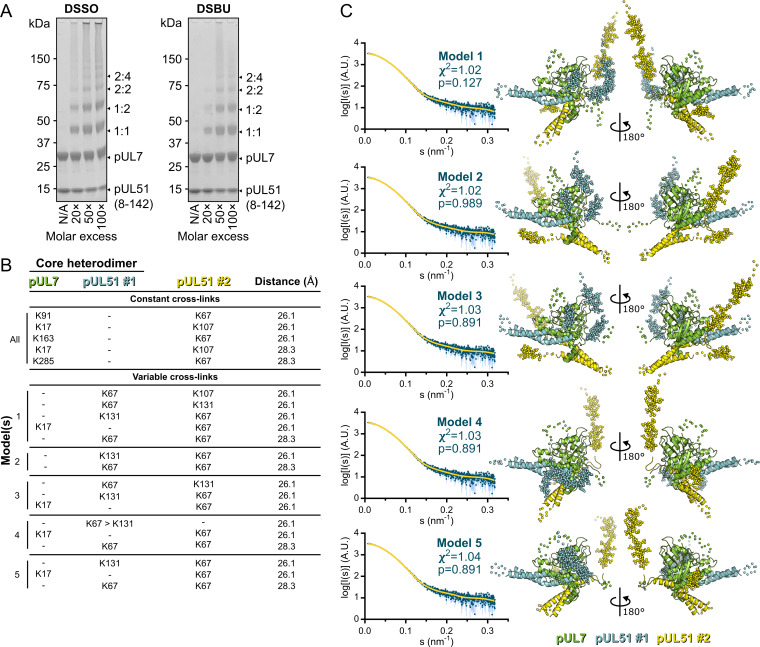
Figure 2. Structure of pUL7 in complex with pUL51.
(A) Hetero-octamer of pUL7 and pUL51(8–142) observed in the crystallographic asymmetric unit. pUL7 and pUL51 are shown as green and cyan ribbons, respectively, in two orthogonal orientations. Inset shows residues arising from the pUL7 cloning tag (pink) that form an eight-stranded β-barrel with residues from pUL51. (B) Core heterodimer of pUL7 (residues 11–296) and pUL51 (residues 41–125). Selected secondary structure elements are labelled. (C) ‘Cut-through’ molecular surface representation of pUL7 (green) showing the intimate interaction interface with the hydrophobic loop and helix α1 of pUL51 (cyan). pUL51 side chains are shown as sticks. (D) Molecular interactions between pUL51 (cyan) and pUL7 (boxed residue names). Hydrophobic and hydrogen bond interactions are in black and red typeface, respectively. (E) Molecular surface representation of pUL7, colored by residue hydrophobicity from white (polar) to orange (hydrophobic). pUL51 is represented as a cyan ribbon with selected side chains shown.