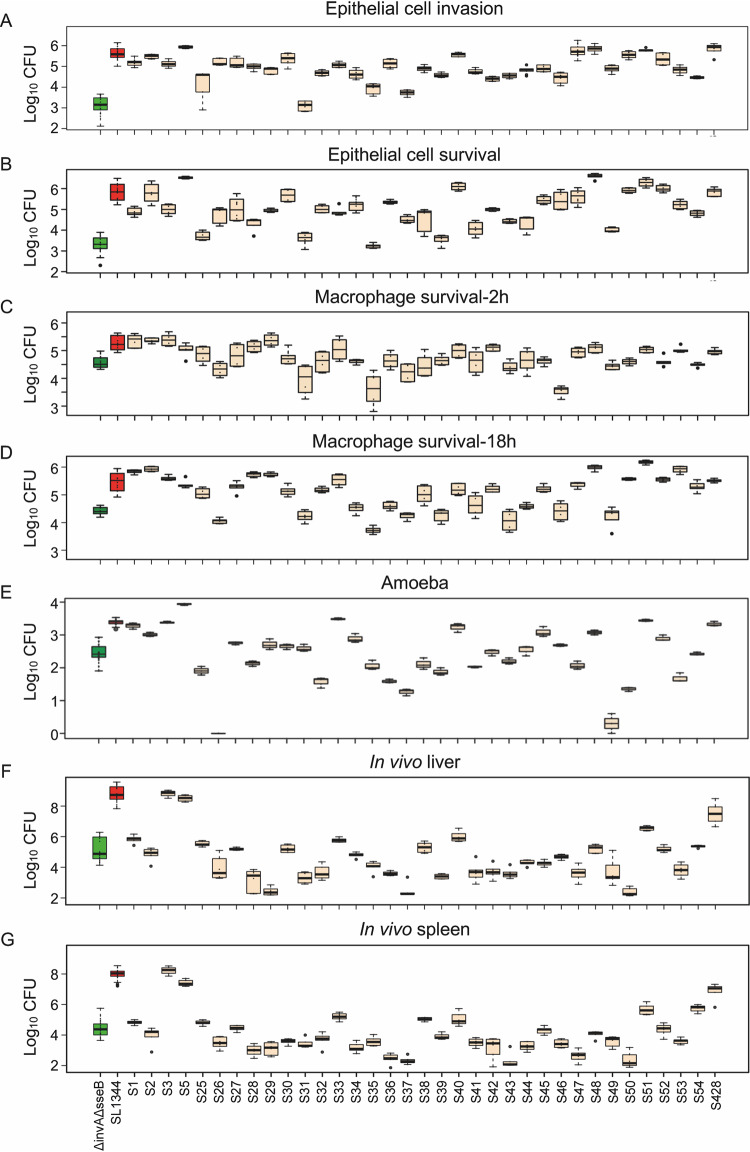
FIG 2.
Phenotyping 35 phylogenetically distant Salmonella serovars in four models of infection. Bacterial load after infection was used as a marker of virulence. Infection of human epithelial cells (A and B), macrophages (C and D), amoebas (E), and in vivo mouse models of systemic disease (F and G). Human epithelial cells were used to measure Salmonella virulence in the form of adhesion, invasion, and replication within epithelial cells. Shown here are invasion (A) measured by intracellular bacterial burden (CFU) of cells 180 min postinfection and survival (B) as measured 18 h postinfection. Human macrophages were used to test strain propensity for phagocytosis and survival measured at 2 (C) and 18 h (D) postinfection. (E) Virulence in the amoeba model was measured by intracellular bacterial burden. In vivo virulence was measured by bacterial burden in mouse liver (F) and spleen (G) 3 days postinfection. Bacterial burden (CFU) in the spleen is shown here. Sample size varied across models and isolates. Either 2, 3, or 4 biological replicates with 3 technical replicates per isolate were used for epithelial cells. Either 2 or 3 biological replicates with 3 technical replicates were used for macrophages. Three biological replicates were performed per isolate in the amoeba and six biological replicates were performed per isolate in the mouse. In all cases, Log10 bacterial load was used as a marker of virulence. The distribution of data for each serovar is shown as a box plot. Low-virulence control ΔinvA ΔsseB strain is shown in green, high-virulence control SL1344 is shown in red, and test strains are in beige.