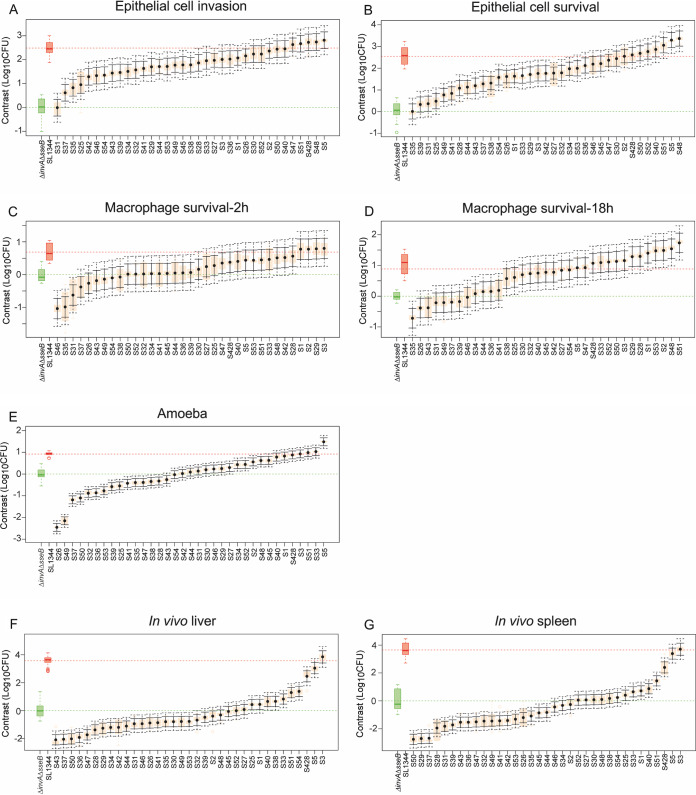
FIG 3.
Distribution of contrasts in the four models of infection for all 35 phylogenetically distant Salmonella serovars. Contrast analysis was used to compare differences in bacterial burden caused by each of the 35 serovars against the low-virulence control ΔinvA ΔsseB strain (mean value shown in green) within all phenotypic traits measured in each model. Epithelial cell invasion (A) and survival (B), macrophage phagocytosis (C) and survival (D), replication in amoeba (E), and bacterial load in liver (F) and spleen (G) of mice are shown. Isolate means are shown by black dots. Orange boxes denote the ranges of values, solid black bars show the 95% confidence intervals, and dotted black bars show the confidence intervals when adjusted for multiple testing using Bonferroni’s correction. All isolates whose adjusted confidence intervals do not cross the green line were significantly different from the ΔinvA ΔsseB strain (P < 0.05). Either 2, 3, or 4 biological replicates with 3 technical replicates per isolate were used for epithelial cells. Either 2 or 3 biological replicates with 3 technical replicates were used for macrophages. Three biological replicates were performed per isolate in the amoeba and six biological replicates were performed per isolate in the mouse.