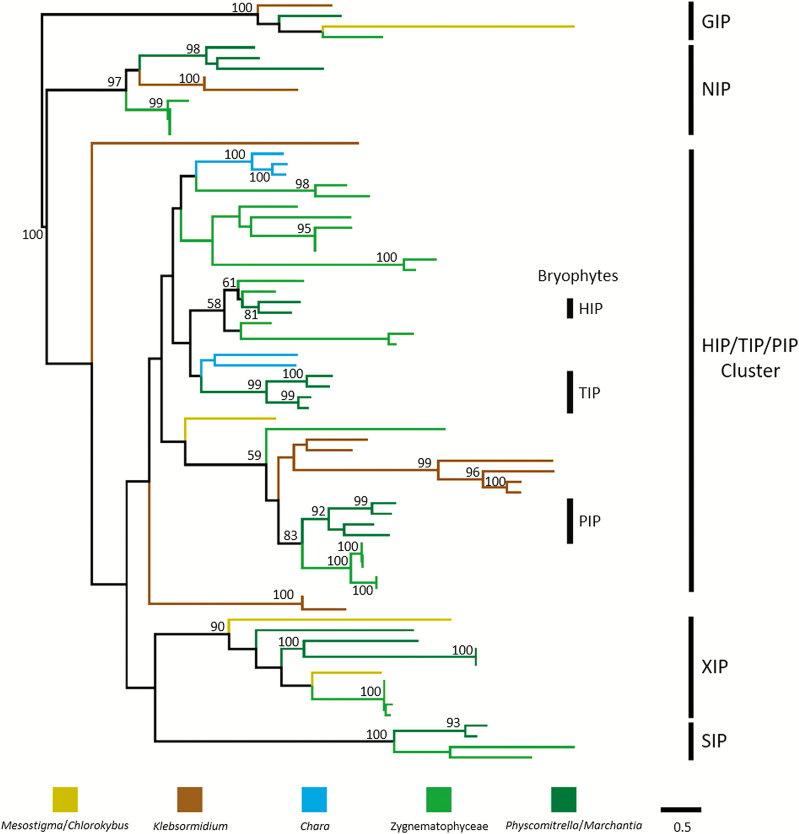
Fig. 4.
Phylogeny of MIPs within streptophytes: 78 protein sequences obtained from seven completed streptophyte genomes were analyzed using the phylogenetic pipeline by ETE3 (www.genome.jp/tools/ete/). See Supplementary Table S1 for a complete list of all proteins aligned. Alignment was performed using Clustal Omega v1.2.1 with the default options (Sievers and Higgins, 2014). The resulting alignment was cleaned using the gappyout algorithm of trimAI v1.4.rev6 (Capella-Gutiérez et al., 2009). The best protein model was selected using NJ tree inference among LG, WAG, JTT, MtREV, Dayhoff, DCMut, RtREV, CpREV, VT, Blosum62, MtMam, MtArt, HIVw, and HIVb models using pmodeltest v1.4. An ML tree was inferred using RAxML v8.1.20 run with model PROTGAMMALGF and default parameters (Stamatakis, 2014). Branch supports were computed out of 100 bootstrapped trees. Boostraps ≥90 are indicated. In addition, a few bootstraps ≥50 within the HIP/TIP/PIP cluster are indicated.