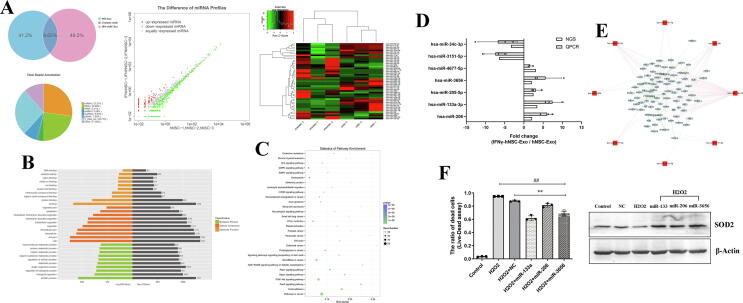
Fig. 6.
NGS and verification of special exosomal miRNAs of IFN-γ-hNSC-Exo. (A) Pie chart representation (Upper left) of the distribution of small RNA categories in exosomes of hNSC-Exo and IFN-γ-hNSC-Exo. Hierarchical clustering (Upper right) and scatter plot (Upper middle) of differentially expressed miRNAs between hNSC-Exo and IFN-γ-hNSC-Exo. Red indicates higher expression; green indicates lower expression. (B) The top 30 GO functions of predicted targets belong to the differentially expressed miRNAs. (C) The top 30 KEGG Pathway enrichment analysis of differential expression genes. (D) Special different exosomal 7 miRNAs (inclusion criteria, |log2(Fold Change)| ≥ 2 and p < 0.01) were validated by qRT-PCR assay, the results were consistent with NGS. (E) The potential targeted genes of special different exosomal 7 miRNAs in IFN-γ-hNSC-Exo were predicted by miRNA-gene network analysis, the hsa-miR-151a-5p was used to contrast. (F) Left: Transfected 3 overexpressed exosomal miRNAs (hsa-miR-206, hsa-miR-133a-3p and hsa-miR-3656) into hNSCs and then treated with H2O2, the proportion of cell survival was increased compared to the H2O2 and NC groups (##p < 0.01 vs·H2O2 group, **p < 0.01 vs. NC group). Right: The over-expression of exosomal miRNAs in cells upregulated anti-oxidative stress SOD2 expression by WB. β-Actin was used as the loading control. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)