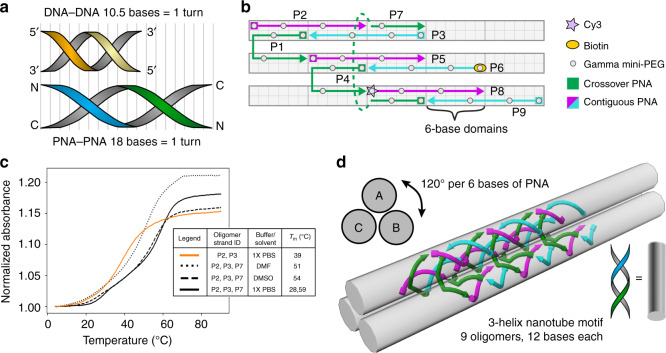
Fig. 1. The design of 3-helical γPNA nanofibers.
a Schematic comparison between a full helical turn of DNA and PNA double helix. B-form DNA is typically stable with 10.5 bases per helical turn while PNA has a helical pitch of 18 bases per turn. b A schematic representation of the structural motif for the SST design that shows 6-base domains and an overall repeat unit of 18 bases. Each PNA oligomer is modified at the 1, 4, and 8 γ-positions with (R)-diethylene glycol (gray dots) to enable pre-organized helical conformation. γPNA oligomers are classified into contiguous (magenta and cyan arrows) or crossover (green arrows) PNAs based on their position in the motif. Specific γPNA oligomers (P8 and P6) are labeled with fluorescent Cy3 (pink star) and biotin (orange oval), respectively, to enable detection of structure formation using fluorescence microscopy. c Melt curve studies show the melting temperatures of 2-oligomer (orange) and 3-oligomer (black) substructures in different solvent conditions vary between 39 and 59 °C. This indicates that 6-base domains are sufficiently stable for γPNA–γPNA systems in PBS (solid curve), DMSO (dashed curves), and DMF (dotted curves). Source data are provided as a Source data file. d 6-Base domains correspond to 120° rise in helical rotation enabling the structural motif to program the assembly of 3-helix nanofibers that can polymerize lengthwise.