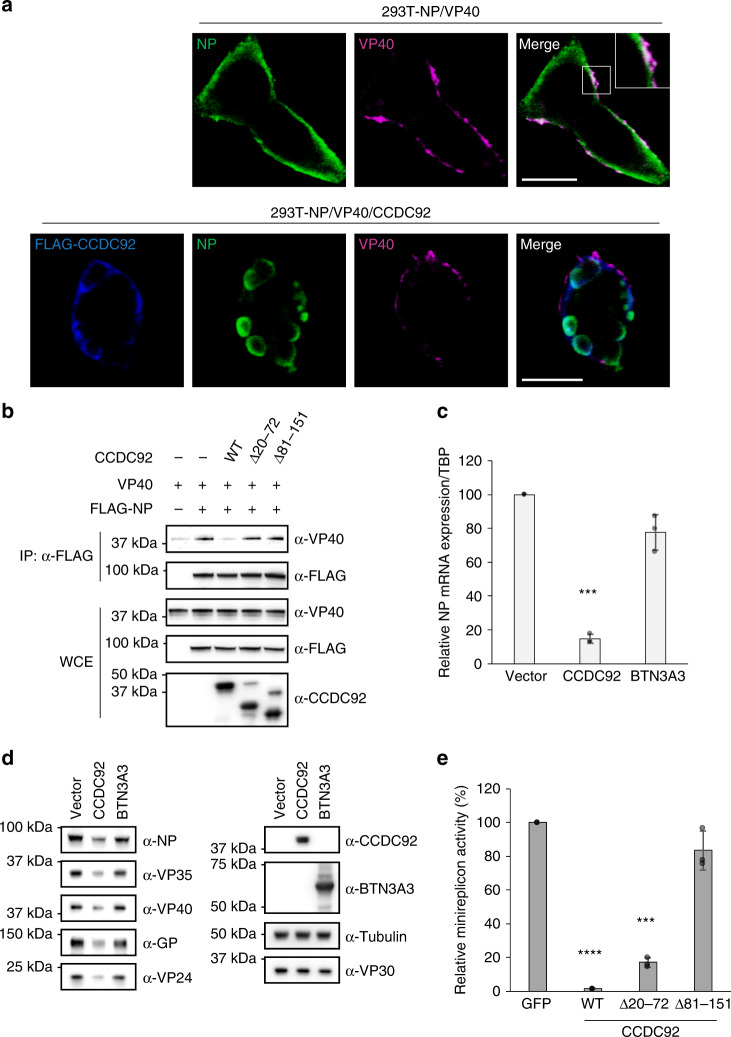
Fig. 5. Inhibition of the NP–VP40 interaction and viral transcription by CCDC92.
a EBOV NP (green) and VP40 (magenta) with or without CCDC92 (blue) in HEK-293T cells transfected with expression vectors for NP and VP40 together with or without FLAG-CCDC92 were visualized with specific antibodies and analyzed by confocal microscopy. The inset is the enlarged image corresponding to the boxed area. Data are representative of two independent experiments. Scale bars, 20 μm. b Interaction between NP and VP40 in the presence of CCDC92 or its deletion mutants in HEK-293T cells transfected with the indicated combination of expression vectors. Cell lysates were immunoprecipitated with anti-FLAG antibody followed by immunoblotting. Data are representative of three independent experiments. IP, immunoprecipitation. WCE, whole-cell extract. c Relative EBOV NP mRNA expression in HEK-293T cells after overexpression of CCDC92 or BTN3A3. Cells were transfected with expression vectors for CCDC92 or BTN3A3, or with an empty control vector for 24 h prior to infection with EBOV∆VP30 at an MOI of 3.0. At 24 h post infection, RNA was extracted and then quantified by qRT-PCR. The results are normalized to TBP mRNA expression, and the data are presented as percentages ± SD (n = 3). (*) indicates a statistically significant difference (p-values of two-tailed Student’s t-tests; ***p < 0.001) from the control. d Cells were transfected and infected as described in c. At 24 h post infection, cells were lysed and the indicated protein expression levels were analyzed by immunoblotting. e Relative luciferase activity in HEK-293T VP30 cells after transfection for 48 h with vectors required for the minireplicon assay, each of the indicated genes, and an internal Renilla luciferase control vector. Data are presented as means ± SD, and are representative of three independent experiments. (*) indicates a statistically significant difference (p-values of two-tailed Student’s t-tests; ***p < 0.001, ****p < 0.0001) from the control. Source data are provided as a Source Data file.