Abstract
Complex multi-cellular organisms are shaped starting from a single-celled zygote, owing to elaborate developmental programs. These programs involve several layers of regulation to orchestrate the establishment of progressively diverging cell type-specific gene expression patterns. In this scenario, epigenetic modifications of chromatin are central in influencing spatiotemporal patterns of gene transcription. In fact, it is generally recognized that epigenetic changes of chromatin states impact on the accessibility of genomic DNA to regulatory proteins. Several lines of evidence highlighted that zebrafish is an excellent vertebrate model for research purposes in the field of developmental epigenetics. In this review, I focus on the dynamic roles recently emerged for histone post-translational modifications (PTMs), histone modifying enzymes, histone variants and histone themselves in the coordination between the precise execution of transcriptional programs and developmental progression in zebrafish. In particular, I first outline a synopsis of the current state of knowledge in this field during early embryogenesis. Then, I present a survey of histone-based epigenetic mechanisms occurring throughout morphogenesis, with a stronger emphasis on cardiac formation. Undoubtedly, the issues addressed in this review take on particular importance in the emerging field of comparative biology of epigenetics, as well as in translational research.
Keywords: histone, histone posttranslational modifications, histone variants, epigenetics, development, maternal-to-zygotic transition, zygotic genome activation, zebrafish
Introduction
The genomic information of eukaryotic cells is confined inside the nucleus in the form of chromatin, a nucleoprotein complex composed primarily of DNA and histone proteins, but also including noncoding RNA and a variety of structural non-histone proteins (Kornberg, 1974; Rodríguez-Campos and Azorín, 2007; Bonev and Cavalli, 2016). The basic repeating unit of this periodic structure, called the “nucleosome core particle,” consists of 147 base pairs of DNA wrapped nearly twice in a left-handed toroidal supercoil around a positively charged protein octamer containing two copies of each of four core histones H2A, H2B, H3, and H4 (Luger et al., 1997; Kornberg and Lorch, 1999). A fifth histone type, H1, interacts with the two internucleosomal linker DNA arms extending from a core particle, thus favoring the establishment of additional hierarchical levels of chromatin compaction (Zhou et al., 2013; Bednar et al., 2017).
Nucleosomes not only act as fundamental units of chromatin packaging, but also play pivotal roles in the coordination between chromatin architecture and functions by means of epigenetic mechanisms (Cavalieri et al., 2009). Among these, covalent post-translational modifications (PTMs) of specific amino acid residues on histones operate in combinatorial fashions either at a single nucleosome level or in a genome-wide manner, thus contributing to an extensive range of biological processes including organism development (Bhaumik et al., 2007; Lee et al., 2010; Cavalieri and Spinelli, 2015). More specifically, the presence of histone PTMs stereochemically alters the binding affinity of the nucleosomes for regulatory complexes that can be recruited or drawn away from chromatin (Smith and Shilatifard, 2010). Although modern molecular biology and mass spectrometry-based methods allowed the discovery of an ever-growing number of histone PTMs, acetylation (ac) and (mono-, di-, and tri-) methylation (me1, me2, and me3, respectively) of lysine (K) residues are the most thoroughly investigated (Di Caro et al., 2007; Zhao and Garcia, 2015; Janssen et al., 2017). Generally speaking, histone PTMs represent repositories of epigenetic memory over multiple generations, especially in those organisms lacking in conventional DNA methylation (Turner, 2009; Cavalieri and Spinelli, 2019). Nonetheless, histone PTMs are not permanent epigenetic marks, because an assorted group of histone-modifying enzymes dynamically governs the attachment or removal of small chemical groups on specific amino acid residues, thereby providing a valuable epigenetic mechanism of cellular adaptation in fluctuating environments (Kouzarides, 2007).
The physicochemical properties of nucleosomes can also be altered by exchanging conventional histone proteins with histone variants showing distinct amino acid sequences compared to their canonical counterparts (Weber and Henikoff, 2014). Such replacements may permit the specific nucleosome recognition, otherwise precluded, by chromatin modifying complexes that will successively appose variant-specific PTMs (Talbert and Henikoff, 2017). Nevertheless, histone variants are often subjected to the same PTMs as their canonical counterparts.
Cell fate decisions made during embryogenesis also depend upon the modulatory control of histone PTMs, histone variants and other epigenetic processes, on the hierarchical cascades of transcriptional events outlining the developmental gene regulatory networks of a given organism (Cavalieri et al., 2008, 2013; Balasubramanian et al., 2019; Horsfield, 2019). In particular, failures in establishing or maintaining proper restrictive and/or permissive patterns of histone PTMs, as well as alterations in histone variant deposition, can seriously disturb the developmental program (Bhaumik et al., 2007; Chen et al., 2013; Maze et al., 2014).
The small freshwater cyprinid Danio rerio, commonly known as zebrafish, offers unique opportunities to investigate histone epigenetic dynamics during vertebrate development. The increasing popularity of this model is due to two main reasons: (1) components of the epigenetic machinery have been widely characterized in zebrafish, showing overall conservation with mammals (Howe et al., 2013; Cavalieri and Spinelli, 2017), and (2) zebrafish embryos are optically translucent and relatively permeable to water-soluble compounds, allowing non-invasive live imaging of morphogenetic events and phenotypes following exposure to environmental stressors acting on the epigenome (Godinho, 2011; Ali et al., 2014). No less important benefits include ease of husbandry and maintenance in laboratory, high fecundity, external fertilization, short life cycle and generation time (Cavalieri and Spinelli, 2017).
Multiple Coordinated Histone-Related Epigenetic Events Accompany Early Embryogenesis
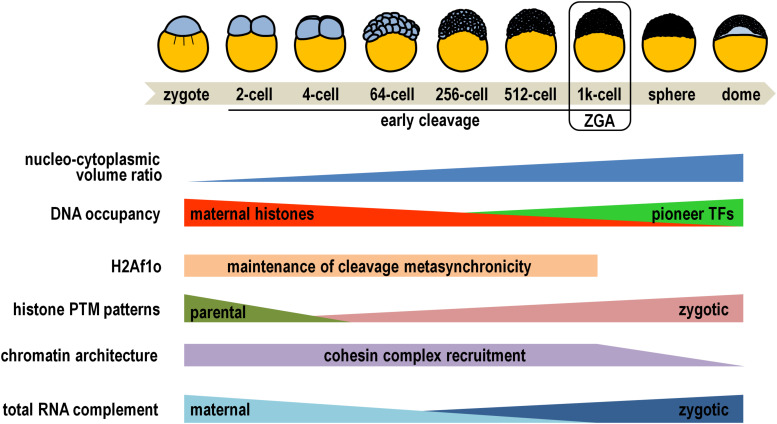
In zebrafish, core histones themselves play a first prominent role during the early cleavage phase of development (Figure 1), when multiple rounds of synchronous cell division convert the newly fertilized egg into a multicellular embryo composed of pluripotent blastomeres (Kimmel et al., 1995). At this time, the nascent embryonic genome is substantially, but not completely, inert in transcription due to dynamic competition for DNA binding between the stoichiometric excess of chromatin-unbound maternal histones and a small set of pioneer transcription factors that, later on, will determine the onset of major zygotic transcription (Lee et al., 2013; Heyn et al., 2014; Joseph et al., 2017). While this competition takes place, the oocyte-specific H2A variant H2Af1o is ubiquitously distributed in the early embryo, where it is critically required for maintaining blastomere cleavage metasynchronicity (Figure 1), probably by conferring a more relatively loose nucleosomal structure than canonical H2A (Kane and Kimmel, 1993; Olivier et al., 2010; Yue et al., 2013).
FIGURE 1.
Diagrammatic representation of key epigenetic changes occurring during early zebrafish embryogenesis. Simplified drawings on the top depict some of the developmental stages, while developmental and epigenetic trends are illustrated below (see main text for details). Please note that placeholder nucleosomes retain parental histone modification patterns throughout early embryogenesis, and that cohesin complex recruitment is restored at 24 h post-fertilization. PTM, post-translational modification; TFs, transcription factors; ZGA, zygotic genome activation.
Within nuclei of cleaving zebrafish embryos, the epigenetic reprogramming process efficiently results in the rapid erasure of the bulk of parental histone PTMs from all nucleosomes except the so called “placeholder” nucleosomes (Figure 1). These specialized nucleosomes harbor the histone variant H2A.Z, termed H2AFV in zebrafish, and are decorated by H3K4me1 (Murphy et al., 2018). These features synergistically attenuate nucleosomal stability and prevent recruitment and/or activity of de novo DNA methyltransferases at promoter of housekeeping and regulatory genes (Lindeman et al., 2011; Hirano et al., 2019). Early developing embryos are loaded with maternal translationally competent mRNAs coding for a wide variety of histone methyltransferases and acetylases, comprehensively accomplishing renewal of zygotic-specific combinations of histone PTMs (Sun et al., 2008; Toyama et al., 2008; Aanes et al., 2011; Lindeman et al., 2011). These epigenetic signatures are overtly detectable at the promoter of about one thousand genes at the 256-cell stage and consist of H3K27ac enrichment restricted almost exclusively to placeholder nucleosomes, and combinations of H3K4me3, H3/H4ac (two PTMs associated with permissive chromatin), H3K27me3 and H3K9me3 (both associated with quiescent chromatin) on canonical nucleosomes (Zhang et al., 2018; Lindeman et al., 2010; Sato et al., 2019). For example, a typical profile foreshadowing high propensity for gene expression comprises co-occurrence of H3K4me3 and H3K9ac/H4ac, while simultaneous accumulation of H3K4me3 and H3K27me3 is reminiscent of bivalent promoters, considered to be in a poised transcriptional state ready for either rapid activation or permanent silencing following the maternal-to-zygotic transition (Puri et al., 2015).
Of not secondary importance, H3K4me3 marks directly correlate with the establishment of well-positioned nucleosome arrays on gene promoters, independently of robust RNA polymerase II binding, and prepare genes for subsequent transcriptional activation (Zhang et al., 2014). In fact, as the nuclear concentration of maternally supplied DNA-unbound histones drops following the progressive increase of the nucleo-cytoplasmic volume ratio at the mid-blastula transition stage (Figure 1), the three pioneer transcription factors Pou5f3, Nanog, and SoxB1, along with the chromatin remodeler Smarca4a, cooperatively finalize chromatin opening by sequential destabilization, displacement, and depletion of nucleosomes occupying the enhancers of developmental genes (Kane and Kimmel, 1993; Haberle et al., 2014; Joseph et al., 2017; Liu et al., 2018; Reisser et al., 2018; Veil et al., 2019).
In the standpoint of the three-dimensional genome architecture, gene-rich accessible chromatin conjointly exhibiting the mentioned permissive histone marks and occupancy of pioneer factors significantly coincides with cohesin complex recruitment (Figure 1), which is a prerequisite for spatial compartmentalization of the zebrafish genome before and during zygotic genome activation (Kaaij et al., 2018; Meier et al., 2018; Vallot and Tachibana, 2020).
Owing to such a strict hierarchical order of epigenetic developmental circuitries, the embryonic genome takes charge of gene expression and the maternal mRNAs are replaced coordinately in all blastomeres by zygotic gene products (Figure 1). Concertedly, the level of H3K4me3 increases further at promoter of transcriptionally active genes, and H3K36me3 (a mark associated with transcription elongation) specifically accumulates on their coding regions (Vastenhouw et al., 2010). By contrast, H3K27me3 enrichment extends dramatically throughout gene body of silenced loci, and heterochromatinization is outlined by removal of permissive PTM marks and concomitant increase in H3K9me3 levels (Lindeman et al., 2011; Laue et al., 2019).
Morphogenesis Completion Engages a Multitude of Histone-Related Epigenetic Activities
Once the maternal-to-zygotic transition has been achieved, the embryo progresses toward gastrulation, during which the blastomere progeny begins to migrate and differentiate to give rise to distinct tissues and organs of the adult fish. Collectively, histone PTMs and their respective modifying enzymes/complexes play pivotal and surprisingly specific roles in ensuring that transcriptional states shift properly from pluripotent to cell type-specific patterns during morphogenesis. In particular, there is now mounting evidence that a multitude of histone lysine methyltransferases (HMTs)/demethylases (KDMs) and acetylases (HATs)/deacetylases (HDACs), as well as peculiar histone variants, are conjointly involved in epigenetic modulation of zebrafish organogenesis, and when their function is impaired, on a case by case basis, the embryo displays several types of organ malformations and/or dysfunctions (Table 1).
TABLE 1.
Overview of studies examining the involvement of key histone modifying enzymes and histone variants in zebrafish development.
| Epigenetic factors | Developmental processes | References | |
| Histone modifiers | CMLO3 | Axis elongation and head formation | Karmodiya et al., 2014 |
| HDAC1 | Craniofacial development, neurogenesis, retinal differentiation, inner hear development, liver, and pancreas morphogenesis | Cunliffe, 2004; Stadler et al., 2005; Yamaguchi et al., 2005; Noël et al., 2008; Zhou et al., 2011; Ignatius et al., 2013; He et al., 2016a | |
| HDAC3 | Liver and posterior lateral line development | Farooq et al., 2008; He et al., 2016b | |
| HDAC4 | Perichondral ossification and pharyngeal skeleton development | DeLaurier et al., 2019 | |
| HDAC5* | Cardiac valve formation | Just et al., 2011 | |
| HDACs | Cardiac valve formation | Kim et al., 2012 | |
| JMJD3 | Myelopoiesis | Yu et al., 2018 | |
| KAT2a and b | Craniofacial development | Sen et al., 2018 | |
| KAT7 | Angiogenesis | Yan et al., 2018 | |
| KDM6ba | Brain, craniofacial, and heart development | Van Laarhoven et al., 2015; Akerberg et al., 2017 | |
| KDM7 | Brain development | Tsukada et al., 2010 | |
| KMT2A | Neurogenesis | Huang et al., 2015 | |
| KMT2D | Brain, craniofacial, and heart development | Van Laarhoven et al., 2015 | |
| LSD1 | Brain development Haematopoiesis | Li et al., 2012; Takeuchi et al., 2015 | |
| MOZ | Pharyngeal segmentation | Miller et al., 2004 | |
| PHF8 | Brain and craniofacial development | Qi et al., 2010 | |
| PRDM3 and 16 | Craniofacial development | Shull et al., 2020 | |
| PRMT1 | Gastrulation movements | Tsai et al., 2011 | |
| PRMT5 | Germline differentiation | Zhu et al., 2019 | |
| PRMT6 | Gastrulation movements | Zhao et al., 2016 | |
| SETDB2 | Gastrulation movements | Du et al., 2014 | |
| SET7/9 | Myoblast differentiation | Tao et al., 2011 | |
| SETD7 | Heart morphogenesis | Kim et al., 2015 | |
| SMYD3 | Cardiac and skeletal muscle development | Fujii et al., 2011; Kim et al., 2015 | |
| SMYD4 | Heart morphogenesis | Xiao et al., 2018 | |
| SMYD5 | Haematopoiesis | Fujii et al., 2016 | |
| Histone variants | H2Af1o | Cell synchrony division before mid-blastula transition | Yue et al., 2013 |
| H2A.FV | Early embryogenesis | Sivasubbu et al., 2006; Madakashira et al., 2017; Murphy et al., 2018 | |
| H2A.Z.2 | Melanocyte differentiation | Raja et al., 2020 | |
| H3.3 | Cranial neural crest differentiation | Cox et al., 2012 | |
| macroH2A1 and 2 | Brain, somite, and fin development | Buschbeck et al., 2009; Gonzalez-Munoz et al., 2019 |
* Inactivation of HDAC5 is required during cardiac valve formation.
As a general rule, several epigenetic modifiers initially showing ubiquitous distribution in the early embryo undergo gradual restriction of their spatial expression pattern as development proceeds. In this scenario, one very pertinent example is provided by the interplay between distinct epigenetic modifications and mechanisms during cardiogenesis. More specifically, the SET and MYND domain-containing SMYD4 HMT becomes progressively restricted to the developing cardiovascular system, where it is directly involved in histone H3K4 di- and tri-methylation, and required to safeguard the proper acetylation level of histone H3 on the same chromatin targets, by recruitment and inactivation of HDAC1 (Xiao et al., 2018). Beyond being an obvious example of cross-talk between chromatin modulators, the SMYD4-HDAC1 association provides an excellent mechanism of tissue-specific regulation of the HDAC1 enzyme, otherwise operating throughout broad sectors of the developing embryo (Cunliffe, 2004; Pillai et al., 2004).
Global levels of H3K4me3 in the chromatin of the forming heart are defined by synergistic involvement of additional HMTs, including SMYD3 and the member of the SET domain-containing family SETD7. In fact, the mono-methyltransferase activity of SETD7 on naïve H3K4 is necessary for successive di- and tri-methylation by SMYD3 (Kim et al., 2015). KMT2D, another component of the SET domain-containing family of HMTs, is non-redundantly implicated in the establishment of H3K4me3 during the progression of cardiac looping (Van Laarhoven et al., 2015). Furthermore, KMT2D associates with KDM6A, a histone demethylase responsible for removal of the H3K27me3 repressive mark from cardiomyocyte chromatin (Issaeva et al., 2007; Van Laarhoven et al., 2015). In this role, the functional cooperation of additional H3K27-specific demethylases, including KDM6Ba and KDM6Bb, is required to promote cardiac trabecular outgrowth (Akerberg et al., 2017). However, this finding does not necessarily mean that H3K27 methylation is completely abolished in cardiomyocytes. Indeed, it should be emphasized that bulk H3K27me3 levels vary widely among different cardiac cell types, suggesting that distinct KDMs could deal with cell type-specific profiling of H3K27me3 through heart morphogenesis (Akerberg et al., 2017).
Although some studies indicated that differentiating cardiac cells necessitate physiological inactivation of specific HDACs, such as HDAC1 and HDAC5, exposure of developing embryos to chemical inhibitors for classes I and II HDACs revealed that general HDAC activity is critically required for the homeostatic balance of histone acetylation in the time window during which heart looping and cardiac valve formation occur (Huynh and McKinsey, 2006; Just et al., 2011; Kim et al., 2012; Xiao et al., 2018).
The combinatorial effect of all the mentioned epigenetic regulators eventually associates with the appropriate expression of cardiac marker genes (Fujii et al., 2011; Just et al., 2011; Kim et al., 2012; Xiao et al., 2018). Of interest, the epigenetic repertoire underlying chromatin of this set of genes typically comprehends elevated occupancy of the replacement histone variant H3.3, as revealed by means of a stable transgenic zebrafish line expressing a biotinylated version of H3.3 exclusively in cardiomyocytes (Goldman et al., 2017). Genome-wide profiling of H3.3-containing nucleosomes also revealed that enrichment of H3.3 alone is a reliable epigenetic indicator of enhancer activity within distinct cardiac subpopulations (Goldman et al., 2017). Conversely, the macroH2A2 histone variant is accumulated throughout the embryo body, in chromatin of both dividing and non-dividing cells, when heart formation processes take place (Buschbeck et al., 2009). It is worth noting, however, that hundreds of genes involved in morphogenesis of cardiac muscle and heart contraction map within chromatin regions enriched in macroH2A2, suggesting important roles for this histone variant (Gonzalez-Munoz et al., 2019). In principle, macroH2A2 occupancy largely coincides with both H3K27me3 and H3K9me3 heterochromatic marks. In spite of this, however, positive and negative mechanistic roles on the degree of chromatin accessibility could be equally postulated for this histone variant, since it is apparently associated with both repressing or activating transcriptional effects, depending on the cell type-specific chromatin context (Gonzalez-Munoz et al., 2019).
Concluding Remarks and Further Perspectives
Cumulative findings argue against the idea that chromatin modifications, especially histone PTMs, could represent an instructive epigenetic code for switching on and off the transcriptional state of genes, as initially thought (Strahl and Allis, 2000; Henikoff and Shilatifard, 2011). Rather, deposition of histone variants and histone PTMs represent dynamic epigenetic features that either modulate chromatin accessibility through recruitment of chromatin remodeling machinery or are added as a consequence of gene transcription (Bartholomew, 2014).
It is clear that histone PTMs and histone variants have fundamental functions throughout zebrafish development, both in early totipotent blastomeres and nearly all differentiating cell types. Whereas the largest proportion of research studies in this field have described genome-wide patterns of histone PTMs and histone variant occupancy, understanding of direct mechanistic relationships between each of these epigenetic marks and specific loci involved in developmental processes is largely missing. On top of that, a lot of histone PTMs characterized in other organisms remain almost unexplored in zebrafish.
Combining characterization of chromatin modification dynamics on the transcriptional outcome of selected genomic regions with detailed phenotypic analysis of developing zebrafish requires advanced in vivo imaging techniques. For example, stated in simple terms, tracking particular types of histone PTMs in living cells requires a probe that specifically recognize them, once established, and a separate tag that allows visual identification of target-probe association. With this rationale, the “mintbody” methodology employs a single-chain variable fragment antibody fused to the enhanced green fluorescent protein to track residue-specific histone PTMs in living organisms, at a single cell level and on specific loci (Yao et al., 2006; Sato et al., 2013; Kimura et al., 2015). Remarkably, generation of viable and fertile transgenic zebrafish expressing a H3K9ac-specific mintbody demonstrated that this molecular tool does not significantly disturb normal cell functions, probably because it binds H3K9ac-containing nucleosomes for intermittent short time intervals, thus allowing regular access to chromatin complexes. Certainly, this powerful imaging analysis will greatly help our understanding of the relative contribution of histone PTMs on cell type-specific gene expression during zebrafish embryogenesis.
Author Contributions
VC conceived the study, obtained the data, and wrote the manuscript.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This research was partially supported by a grant from the University of Palermo (Fondo Finalizzato Ricerca, grant number FFR-D15-160962) to VC.
References
- Aanes H., Winata C. L., Lin C. H., Chen J. P., Srinivasan K. G., Lee S. G., et al. (2011). Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res. 21 1328–1338. 10.1101/gr.116012.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerberg A. A., Henner A., Stewart S., Stankunas K. (2017). Histone demethylases Kdm6ba and Kdm6bb redundantly promote cardiomyocyte proliferation during zebrafish heart ventricle maturation. Dev. Biol. 426 84–96. 10.1016/j.ydbio.2017.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S., Aalders J., Richardson M. K. (2014). Teratological effects of a panel of sixty water-soluble toxicants on zebrafish development. Zebrafish 11 129–141. 10.1089/zeb.2013.0901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S., Raghunath A., Perumal E. (2019). Role of epigenetics in zebrafish development. Gene 718:144049. 10.1016/j.gene.2019.144049 [DOI] [PubMed] [Google Scholar]
- Bartholomew B. (2014). Regulating the chromatin landscape: structural and mechanistic perspectives. Annu. Rev. Biochem. 83 671–696. 10.1146/annurev-biochem-051810-093157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar J., Garcia-Saez I., Boopathi R., Cutter A. R., Papai G., Reymer A., et al. (2017). Structure and dynamics of a 197 bp nucleosome in complex with linker histone H1. Mol. Cell. 66 384–397.e8. 10.1016/j.molcel.2017.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S. R., Smith E., Shilatifard A. (2007). Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 14 1008–1016. 10.1038/nsmb1337 [DOI] [PubMed] [Google Scholar]
- Bonev B., Cavalli G. (2016). Organization and function of the 3D genome. Nat. Rev. Genet. 17 661–678. 10.1038/nrg.2016.112 [DOI] [PubMed] [Google Scholar]
- Buschbeck M., Uribesalgo I., Wibowo I., Rué P., Martin D., Gutierrez A., et al. (2009). The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat. Struct. Mol. Biol. 16 1074–1079. 10.1038/nsmb.1665 [DOI] [PubMed] [Google Scholar]
- Cavalieri V., Di Bernardo M., Anello L., Spinelli G. (2008). cis-Regulatory sequences driving the expression of the Hbox12 homeobox-containing gene in the presumptive aboral ectoderm territory of the Paracentrotus lividus sea urchin embryo. Dev. Biol. 321 455–469. 10.1016/j.ydbio.2008.06.006 [DOI] [PubMed] [Google Scholar]
- Cavalieri V., Melfi R., Spinelli G. (2009). Promoter activity of the sea urchin (Paracentrotus lividus) nucleosomal H3 and H2A and linker H1 {alpha}-histone genes is modulated by enhancer and chromatin insulator. Nucleic Acids Res. 37 7407–7415. 10.1093/nar/gkp859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri V., Melfi R., Spinelli G. (2013). The Compass-like locus, exclusive to the Ambulacrarians, encodes a chromatin insulator binding protein in the sea urchin embryo. PLoS Genet. 9:e1003847. 10.1371/journal.pgen.1003847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri V., Spinelli G. (2015). Ectopic hbox12 expression evoked by histone deacetylase inhibition disrupts axial specification of the sea urchin embryo. PLoS One 10:e0143860. 10.1371/journal.pone.0143860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri V., Spinelli G. (2017). Environmental epigenetics in zebrafish. Epigenet. Chrom. 10:46. 10.1186/s13072-017-0154-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri V., Spinelli G. (2019). “Histone-mediated transgenerational epigenetics,” in Translational Epigenetics (Second Edition), ed. Tollefsbol T. O. (Amsterdam: Elsevier; ), 157–183. 10.1016/B978-0-12-816363-4.00008-0 [DOI] [Google Scholar]
- Chen P., Zhao J., Li G. (2013). Histone variants in development and diseases. J. Genet. Genomics 40 355–365. 10.1016/j.jgg.2013.05.001 [DOI] [PubMed] [Google Scholar]
- Cox S. G., Kim H., Garnett A. T., Medeiros D. M., An W., Crump J. G. (2012). An essential role of variant histone H3.3 for ectomesenchyme potential of the cranial neural crest. PLoS Genet. 8:e1002938. 10.1371/journal.pgen.1002938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe V. T. (2004). Histone deacetylase 1 is required to repress notch target gene expression during zebrafish neurogenesis and to maintain the production of motoneurones in response to hedgehog signalling. Development 131 2983–2995. 10.1242/dev.01166 [DOI] [PubMed] [Google Scholar]
- DeLaurier A., Alvarez C. L., Wiggins K. J. (2019). hdac4 mediates perichondral ossification and pharyngeal skeleton development in the zebrafish. PeerJ 7:e6167. 10.7717/peerj.6167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Caro V., Cavalieri V., Melfi R., Spinelli G. (2007). Constitutive promoter occupancy by the MBF-1 activator and chromatin modification of the developmental regulated sea urchin alpha-H2A histone gene. J. Mol. Biol. 365 1285–1297. 10.1016/j.jmb.2006.10.098 [DOI] [PubMed] [Google Scholar]
- Du T. T., Xu P. F., Dong Z. W., Fan H. B., Jin Y., Dong M., et al. (2014). Setdb2 controls convergence and extension movements during zebrafish gastrulation by transcriptional regulation of dvr1. Dev. Biol. 392 233–244. 10.1016/j.ydbio.2014.05.022 [DOI] [PubMed] [Google Scholar]
- Farooq M., Sulochana K. N., Pan X., To J., Sheng D., Gong Z., et al. (2008). Histone deacetylase 3 (hdac3) is specifically required for liver development in zebrafish. Dev. Biol. 317 336–353. 10.1016/j.ydbio.2008.02.034 [DOI] [PubMed] [Google Scholar]
- Fujii T., Tsunesumi S., Sagara H., Munakata M., Hisaki Y., Sekiya T., et al. (2016). Smyd5 plays pivotal roles in both primitive and definitive hematopoiesis during zebrafish embryogenesis. Sci. Rep. 6:29157. 10.1038/srep29157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T., Tsunesumi S., Yamaguchi K., Watanabe S., Furukawa Y. (2011). Smyd3 is required for the development of cardiac and skeletal muscle in zebrafish. PLoS One 6:e23491. 10.1371/journal.pone.0023491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho L. (2011). Live imaging of zebrafish development. Cold Spring Harb. Protoc. 2011 770–777. 10.1101/pdb.top119 [DOI] [PubMed] [Google Scholar]
- Goldman J. A., Kuzu G., Lee N., Karasik J., Gemberling M., Foglia M. J., et al. (2017). Resolving heart regeneration by replacement histone profiling. Dev. Cell. 40 392–404.e5. 10.1016/j.devcel.2017.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Munoz E., Arboleda-Estudillo Y., Chanumolu S. K., Otu H. H., Cibelli J. B. (2019). Zebrafish macroH2A variants have distinct embryo localization and function. Sci. Rep. 9:8632. 10.1038/s41598-019-45058-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V., Li N., Hadzhiev Y., Plessy C., Previti C., Nepal C., et al. (2014). Two independent transcription initiation codes overlap on vertebrate core promoters. Nature 507 381–385. 10.1038/nature12974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Tang D., Li W., Chai R., Li H. (2016a). Histone deacetylase 1 is required for the development of the zebrafish inner ear. Sci. Rep. 6:16535. 10.1038/srep16535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Wang Z., Sun S., Tang D., Li W., Chai R., et al. (2016b). HDAC3 is required for posterior lateral line development in Zebrafish. Mol. Neurobiol. 53 5103–5117. 10.1007/s12035-015-9433-6 [DOI] [PubMed] [Google Scholar]
- Henikoff S., Shilatifard A. (2011). Histone modification: cause or cog? Trends Genet. 27 389–396. 10.1016/j.tig.2011.06.006 [DOI] [PubMed] [Google Scholar]
- Heyn P., Kircher M., Dahl A., Kelso J., Tomancak P., Kalinka A. T., et al. (2014). The earliest transcribed zygotic genes are short, newly evolved, and different across species. Cell Rep. 6 285–292. 10.1016/j.celrep.2013.12.030 [DOI] [PubMed] [Google Scholar]
- Hirano R., Kujirai T., Negishi L., Kurumizaka H. (2019). Biochemical characterization of the placeholder nucleosome for DNA hypomethylation maintenance. Biochem. Biophys. Rep. 18:100634. 10.1016/j.bbrep.2019.100634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield J. A. (2019). Packaging development: how chromatin controls transcription in zebrafish embryogenesis. Biochem. Soc. Trans. 47 713–724. 10.1042/BST20180617 [DOI] [PubMed] [Google Scholar]
- Howe K., Clark M. D., Torroja C. F., Torrance J., Berthelot C., Muffato M., et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496 498–503. 10.1038/nature12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. C., Shih H. Y., Lin S. J., Chiu C. C., Ma T. L., Yeh T. H., et al. (2015). The epigenetic factor Kmt2a/Mll1 regulates neural progenitor proliferation and neuronal and glial differentiation. Dev. Neurobiol. 75 452–462. 10.1002/dneu.22235 [DOI] [PubMed] [Google Scholar]
- Huynh Q. K., McKinsey T. A. (2006). Protein kinase D directly phosphorylates histone deacetylase 5 via a random sequential kinetic mechanism. Arch. Biochem. Biophys. 450 141–148. 10.1016/j.abb.2006.02.014 [DOI] [PubMed] [Google Scholar]
- Ignatius M. S., Unal Eroglu A., Malireddy S., Gallagher G., Nambiar R. M., Henion P. D. (2013). Distinct functional and temporal requirements for zebrafish Hdac1 during neural crest-derived craniofacial and peripheral neuron development. PLoS One 8:e63218. 10.1371/journal.pone.0063218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaeva I., Zonis Y., Rozovskaia T., Orlovsky K., Croce C. M., Nakamura T., et al. (2007). Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell. Biol. 27 1889–1903. 10.1128/MCB.01506-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen K. A., Sidoli S., Garcia B. A. (2017). Recent achievements in characterizing the histone code and approaches to integrating epigenomics and systems biology. Methods Enzymol. 586 359–378. 10.1016/bs.mie.2016.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S. R., Pálfy M., Hilbert L., Kumar M., Karschau J., Zaburdaev V., et al. (2017). Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. eLife 6:e23326. 10.7554/eLife.23326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just S., Berger I. M., Meder B., Backs J., Keller A., Marquart S., et al. (2011). Protein kinase D2 controls cardiac valve formation in zebrafish by regulating histone deacetylase 5 activity. Circulation 124 324–334. 10.1161/CIRCULATIONAHA.110.003301 [DOI] [PubMed] [Google Scholar]
- Kaaij L. J. T., van der Weide R. H., Ketting R. F., de Wit E. (2018). Systemic loss and gain of chromatin architecture throughout zebrafish development. Cell Rep. 24 1–10.e8. 10.1016/j.celrep.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane D. A., Kimmel C. B. (1993). The zebrafish midblastula transition. Development 119 447–456. [DOI] [PubMed] [Google Scholar]
- Karmodiya K., Anamika K., Muley V., Pradhan S. J., Bhide Y., Galande S. (2014). Camello, a novel family of Histone Acetyltransferases that acetylate histone H4 and is essential for zebrafish development. Sci. Rep. 4:6076. 10.1038/srep06076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. D., Kim E., Koun S., Ham H. J., Rhee M., Kim M. J., et al. (2015). Proper activity of histone H3 Lysine 4 (H3K4) methyltransferase is required for morphogenesis during zebrafish cardiogenesis. Mol. Cells 38 580–586. 10.14348/molcells.2015.0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. S., Kim M. J., Koo T. H., Kim J. D., Koun S., Ham H. J., et al. (2012). Histone deacetylase is required for the activation of Wnt/β-catenin signaling crucial for heart valve formation in zebrafish embryos. Biochem. Biophys. Res. Commun. 423 140–146. 10.1016/j.bbrc.2012.05.098 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203 253–310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kimura H., Hayashi-Takanaka Y., Stasevich T. J., Sato Y. (2015). Visualizing posttranslational and epigenetic modifications of endogenous proteins in vivo. Histochem. Cell. Biol. 144 101–109. 10.1007/s00418-015-1344-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D. (1974). Chromatin structure: a repeating unit of histones and DNA. Science 184 868–871. 10.1126/science.184.4139.868 [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Lorch Y. (1999). Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98 285–294. 10.1016/s0092-8674(00)81958-3 [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Laue K., Rajshekar S., Courtney A. J., Lewis Z. A., Goll M. G. (2019). The maternal to zygotic transition regulates genome-wide heterochromatin establishment in the zebrafish embryo. Nat. Commun. 10:1551. 10.1038/s41467-019-09582-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Smith E., Shilatifard A. (2010). The language of histone crosstalk. Cell 142 682–685. 10.1016/j.cell.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. T., Bonneau A. R., Takacs C. M., Bazzini A. A., DiVito K. R., Fleming E. S., et al. (2013). Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503 360–364. 10.1038/nature12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Sun Y., Dou C., Chen J., Zhang J. (2012). Lysine-specific demethylase 1 expression in zebrafish during the early stages of neuronal development. Neural Regen. Res. 7:2719. 10.3969/j.issn.1673-5374.2012.34.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman L. C., Andersen I. S., Reiner A. H., Li N., Aanes H., Østrup O., et al. (2011). Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev. Cell. 21 993–1004. 10.1016/j.devcel.2011.10.008 [DOI] [PubMed] [Google Scholar]
- Lindeman L. C., Winata C. L., Aanes H., Mathavan S., Alestrom P., Collas P. (2010). Chromatin states of developmentally-regulated genes revealed by DNA and histone methylation patterns in zebrafish embryos. Int. J. Dev. Biol. 54 803–813. 10.1387/ijdb.103081ll [DOI] [PubMed] [Google Scholar]
- Liu G., Wang W., Hu S., Wang X., Zhang Y. (2018). Inherited DNA methylation primes the establishment of accessible chromatin during genome activation. Genome Res. 28 998–1007. 10.1101/gr.228833.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389 251–260. 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- Madakashira B., Corbett L., Zhang C., Paoli P., Casement J. W., Mann J., et al. (2017). Variant Histone H2afv reprograms DNA methylation during early zebrafish development. Epigenetics 12 811–824. 10.1080/15592294.2017.1359382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I., Noh K. M., Soshnev A. A., Allis C. D. (2014). Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat. Rev. Genet. 15 259–271. 10.1038/nrg3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier M., Grant J., Dowdle A., Thomas A., Gerton J., Collas P., et al. (2018). Cohesin facilitates zygotic genome activation in zebrafish. Development 145:dev156521. 10.1242/dev.156521 [DOI] [PubMed] [Google Scholar]
- Miller C. T., Maves L., Kimmel C. B. (2004). Moz regulates Hox expression and pharyngeal segmental identity in zebrafish. Development 131 2443–2461. 10.1242/dev.01134 [DOI] [PubMed] [Google Scholar]
- Murphy P. J., Wu S. F., James C. R., Wike C. L., Cairns B. R. (2018). Placeholder nucleosomes underlie germline-to-embryo DNA methylation reprogramming. Cell 172 993–1006.e13. 10.1016/j.cell.2018.01.022 [DOI] [PubMed] [Google Scholar]
- Noël E. S., Casal-Sueiro A., Busch-Nentwich E., Verkade H., Dong P. D., Stemple D. L., et al. (2008). Organ-specific requirements for Hdac1 in liver and pancreas formation. Dev. Biol. 322 237–250. 10.1016/j.ydbio.2008.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier N., Luengo-Oroz M. A., Duloquin L., Faure E., Savy T., Veilleux I., et al. (2010). Cell lineage reconstruction of early zebrafish embryos using label-free nonlinear microscopy. Science 329 967–971. 10.1126/science.1189428 [DOI] [PubMed] [Google Scholar]
- Pillai R., Coverdale L. E., Dubey G., Martin C. C. (2004). Histone deacetylase 1 (HDAC-1) required for the normal formation of craniofacial cartilage and pectoral fins of the zebrafish. Dev. Dyn. 231 647–654. 10.1002/dvdy.20168 [DOI] [PubMed] [Google Scholar]
- Puri D., Gala H., Mishra R., Dhawan J. (2015). High-wire act: the poised genome and cellular memory. FEBS J. 282 1675–1691. 10.1111/febs.13165 [DOI] [PubMed] [Google Scholar]
- Qi H. H., Sarkissian M., Hu G. Q., Wang Z., Bhattacharjee A., Gordon D. B., et al. (2010). Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466 503–507. 10.1038/nature09261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja D. A., Subramaniam Y., Aggarwal A., Gotherwal V., Babu A., Tanwar J., et al. (2020). Histone variant dictates fate biasing of neural crest cells to melanocyte lineage. Development 147:dev182576. 10.1242/dev.182576 [DOI] [PubMed] [Google Scholar]
- Reisser M., Palmer A., Popp A. P., Jahn C., Weidinger G., Gebhardt J. C. M. (2018). Single-molecule imaging correlates decreasing nuclear volume with increasing TF-chromatin associations during zebrafish development. Nat. Commun. 9:5218. 10.1038/s41467-018-07731-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Campos A., Azorín F. (2007). RNA is an integral component of chromatin that contributes to its structural organization. PLoS One 2:e1182. 10.1371/journal.pone.0001182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Hilbert L., Oda H., Wan Y., Heddleston J. M., Chew T. L., et al. (2019). Histone H3K27 acetylation precedes active transcription during zebrafish zygotic genome activation as revealed by live-cell analysis. Development 146:dev179127. 10.1242/dev.179127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Mukai M., Ueda J., Muraki M., Stasevich T. J., Horikoshi N., et al. (2013). Genetically encoded system to track histone modification in vivo. Sci. Rep. 3:2436. 10.1038/srep02436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R., Pezoa S. A., Carpio Shull L., Hernandez-Lagunas L., Niswander L. A., Artinger K. B. (2018). Kat2a and Kat2b acetyltransferase activity regulates craniofacial cartilage and bone differentiation in zebrafish and mice. J. Dev. Biol. 6:27. 10.3390/jdb6040027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull L. C., Sen R., Menzel J., Goyama S., Kurokawa M., Artinger K. B. (2020). The conserved and divergent roles of Prdm3 and Prdm16 in zebrafish and mouse craniofacial development. Dev. Biol. 461 132–144. 10.1016/j.ydbio.2020.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasubbu S., Balciunas D., Davidson A. E., Pickart M. A., Hermanson S. B., Wangensteen K. J., et al. (2006). Gene-breaking transposon mutagenesis reveals an essential role for histone H2afza in zebrafish larval development. Mech. Dev. 123 513–529. 10.1016/j.mod.2006.06.002 [DOI] [PubMed] [Google Scholar]
- Smith E., Shilatifard A. (2010). The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol. Cell 40 689–701. 10.1016/j.molcel.2010.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler J. A., Shkumatava A., Norton W. H., Rau M. J., Geisler R., Fischer S., et al. (2005). Histone deacetylase 1 is required for cell cycle exit and differentiation in the zebrafish retina. Dev. Dyn. 233 883–889. 10.1002/dvdy.20427 [DOI] [PubMed] [Google Scholar]
- Strahl B. D., Allis C. D. (2000). The language of covalent histone modifications. Nature 403 41–45. 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- Sun X. J., Xu P. F., Zhou T., Hu M., Fu C. T., Zhang Y., et al. (2008). Genome-wide survey and developmental expression mapping of zebrafish SET domain-containing genes. PLoS One 3:e1499. 10.1371/journal.pone.0001499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M., Fuse Y., Watanabe M., Andrea C. S., Takeuchi M., Nakajima H., et al. (2015). LSD1/KDM1A promotes hematopoietic commitment of hemangioblasts through downregulation of Etv2. Proc. Natl. Acad. Sci. U.S.A. 112 13922–13927. 10.1073/pnas.1517326112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P. B., Henikoff S. (2017). Histone variants on the move: substrates for chromatin dynamics. Nat. Rev. Mol. Cell. Biol. 18 115–126. 10.1038/nrm.2016.148 [DOI] [PubMed] [Google Scholar]
- Tao Y., Neppl R. L., Huang Z. P., Chen J., Tang R. H., Cao R., et al. (2011). The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J. Cell. Biol. 194 551–565. 10.1083/jcb.201010090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama R., Rebbert M. L., Dey A., Ozato K., Dawid I. B. (2008). Brd4 associates with mitotic chromosomes throughout early zebrafish embryogenesis. Dev. Dyn. 237 1636–1644. 10.1002/dvdy.21576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. J., Pan H., Hung C. M., Hou P. T., Li Y. C., Lee Y. J., et al. (2011). The predominant protein arginine methyltransferase PRMT1 is critical for zebrafish convergence and extension during gastrulation. FEBS J. 278 905–917. 10.1111/j.1742-4658.2011.08006.x [DOI] [PubMed] [Google Scholar]
- Tsukada Y. I., Ishitani T., Nakayama K. I. (2010). KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 24 432–437. 10.1101/gad.1864410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B. M. (2009). Epigenetic responses to environmental change and their evolutionary implications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364 3403–3418. 10.1098/rstb.2009.0125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallot A., Tachibana K. (2020). The emergence of genome architecture and zygotic genome activation. Curr. Opin. Cell. Biol. 64 50–57. 10.1016/j.ceb.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laarhoven P. M., Neitzel L. R., Quintana A. M., Geiger E. A., Zackai E. H., Clouthier D. E., et al. (2015). Kabuki syndrome genes KMT2D and KDM6A: functional analyses demonstrate critical roles in craniofacial, heart and brain development. Hum. Mol. Genet. 24 4443–4453. 10.1093/hmg/ddv180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw N. L., Zhang Y., Woods I. G., Imam F., Regev A., Liu X. S., et al. (2010). Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464 922–926. 10.1038/nature08866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veil M., Yampolsky L. Y., Grüning B., Onichtchouk D. (2019). Pou5f3, SoxB1, and Nanog remodel chromatin on high nucleosome affinity regions at zygotic genome activation. Genome Res. 29 383–395. 10.1101/gr.240572.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C. M., Henikoff S. (2014). Histone variants: dynamic punctuation in transcription. Genes Dev. 28 672–682. 10.1101/gad.238873.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao D., Wang H., Hao L., Guo X., Ma X., Qian Y., et al. (2018). The roles of SMYD4 in epigenetic regulation of cardiac development in zebrafish. PLoS Genet. 14:e1007578. 10.1371/journal.pgen.1007578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Tonou-Fujimori N., Komori A., Maeda R., Nojima Y., Li H., et al. (2005). Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing Wnt and Notch signaling pathways. Development 132 3027–3043. 10.1242/dev.01881 [DOI] [PubMed] [Google Scholar]
- Yan M. S., Turgeon P. J., Man H. J., Dubinsky M. K., Ho J. J. D., El-Rass S., et al. (2018). Histone acetyltransferase 7 (KAT7)-dependent intragenic histone acetylation regulates endothelial cell gene regulation. J. Biol. Chem. 293 4381–4402. 10.1074/jbc.RA117.001383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Munson K. M., Webb W. W., Lis J. T. (2006). Dynamics of heat shock factor association with native gene loci in living cells. Nature 442 1050–1053. 10.1038/nature05025 [DOI] [PubMed] [Google Scholar]
- Yu S. H., Zhu K. Y., Zhang F., Wang J., Yuan H., Chen Y., et al. (2018). The histone demethylase Jmjd3 regulates zebrafish myeloid development by promoting spi1 expression. Biochim. Biophys. Acta Gene Regul. Mech. 1861 106–116. 10.1016/j.bbagrm.2017.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue H. M., Li Z., Wu N., Liu Z., Wang Y., Gui J. F. (2013). Oocyte-specific H2A variant H2af1o is required for cell synchrony before midblastula transition in early zebrafish embryos. Biol. Reprod. 89:82. 10.1095/biolreprod.113.108043 [DOI] [PubMed] [Google Scholar]
- Zhang B., Wu X., Zhang W., Shen W., Sun Q., Liu K., et al. (2018). Widespread enhancer dememorization and promoter priming during parental-to-zygotic transition. Mol. Cell. 72 673–686.e6. 10.1016/j.molcel.2018.10.017 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Vastenhouw N. L., Feng J., Fu K., Wang C., Ge Y., et al. (2014). Canonical nucleosome organization at promoters forms during genome activation. Genome Res. 24 260–266. 10.1101/gr.157750.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. X., Zhang Y. B., Ni P. L., Wu Z. L., Yan Y. C., Li Y. P. (2016). Protein arginine methyltransferase 6 (Prmt6) is essential for early zebrafish development through the direct suppression of gadd45αa stress sensor gene. J. Biol. Chem. 291 402–412. 10.1074/jbc.M115.666347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Garcia B. A. (2015). Comprehensive catalog of currently documented histone modifications. Cold Spring Harb. Perspect. Biol. 7:a025064. 10.1101/cshperspect.a025064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B. R., Feng H., Kato H., Dai L., Yang Y., Zhou Y., et al. (2013). Structural insights into the histone H1-nucleosome complex. Proc. Natl. Acad. Sci. U.S.A. 110 19390–19395. 10.1073/pnas.1314905110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Liang I. C., Yee N. S. (2011). Histone deacetylase 1 is required for exocrine pancreatic epithelial proliferation in development and cancer. Cancer Biol. Ther. 11 659–670. 10.4161/cbt.11.7.14720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Zhang D., Liu X., Yu G., Cai X., Xu C., et al. (2019). Zebrafish prmt5 arginine methyltransferase is essential for germ cell development. Development 146:dev179572. 10.1242/dev.179572 [DOI] [PubMed] [Google Scholar]