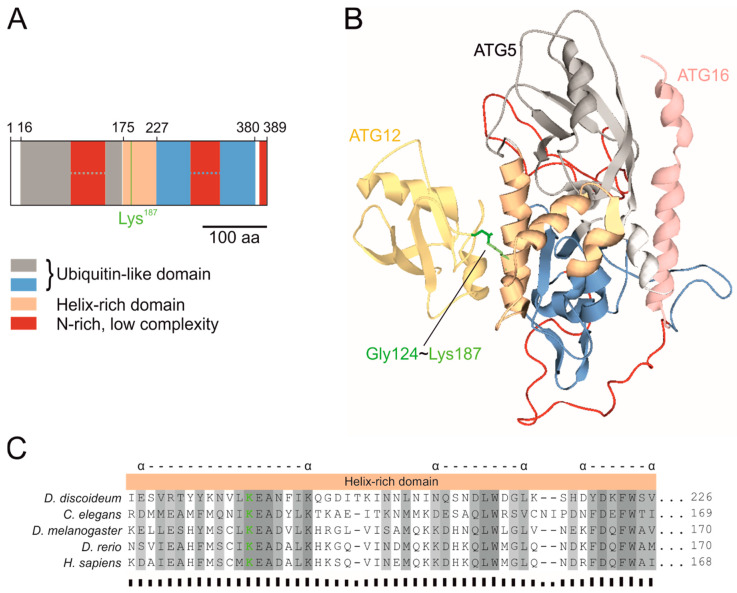
Figure 2.
ATG5 domain composition, 3D structure and multiple sequence alignment of the helix-rich domain (HRD). (A) Schematic representation of the predicted ATG5 domains. Two ubiquitin-like domains (UblDs) flank the HRD, which contains Lys187 (green line), that forms the covalent bond with the C-terminal glycine of ATG12. Both UblDs are interrupted by N-rich, low-complexity regions. The domains and regions were predicted by InterPro [57] and SMART [56], respectively. (B) Predicted 3D structure of the D. discoideum ATG12~5/16N complex as ribbon diagram by homology modelling [58] using the human complex as template (PDB ID: 4gdk) [60]. ATG12 is shown in yellow, the ATG16N in rose, and for ATG5 the same colors as in (A) were used, to emphasise the domains and regions. The amino acid side chain of ATG5 Lys187 is shown in green. (C) Multiple sequence alignment of the conserved HRD of ATG5 orthologues from different organisms. The multiple sequence alignment was performed with Clustal Omega [54]. Amino acid residues are numbered on the right and sequence similarity is indicated by shading. Dark grey represents identical amino acid residues, medium grey highlights amino acids with very similar properties (roughly equivalent to > 0.5 scoring in the Gonnet PAM 250 matrix) and light grey amino acids with slightly similar properties (roughly equivalent to scoring ≤ 0.5 and > 0 in the Gonnet PAM 250 matrix). Sequence conservation is indicated below the alignment by bar sizes [55]. Alpha helices as predicted in the homology model for D. discoideum ATG5 in (B) are marked with α above the alignment. The complete sequence alignment of ATG5 orthologs is shown in Figure S2.