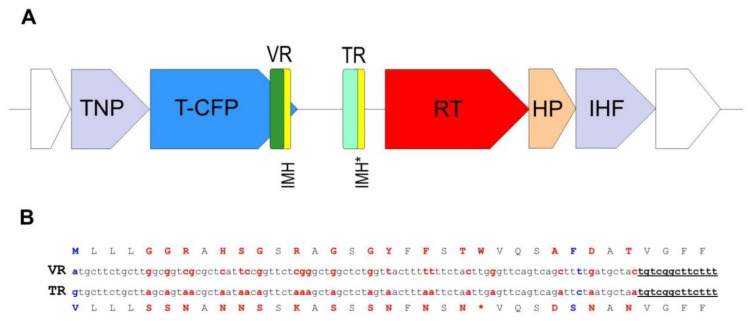
Figure 3.
Schematic structure of the crAssphage LMMB diversity generating retroelement (DGR). (A) DGR in the genome of the crAssphage LMMB was verified using myDGR software (https://omics.informatics.indiana.edu/myDGR). The target gene (T-CFP) is marked with blue, the gene encoding reverse transcriptase (RT) is marked with red, the variable repeat (VR) is marked with green, the target repeat (TR) is marked with light green, both IMH and IMH* are marked with yellow. The flanking genes, encoding a tail-needle protein (TNP) and integration host factor (IHF) are marked with purple, the hypothetical protein (HP) is marked with orange. (B) Comparison of the VR and TR of the crAssphage LMMB shown for the DNA and aa sequences (The TR is not translated in vivo.). Variable positions in the VR and the corresponding adenine residues in the TR are marked with red letters. Non A-to-N mutations are indicated with blue letters. The IMH and IMH* sequences are underlined.