Figure 1.
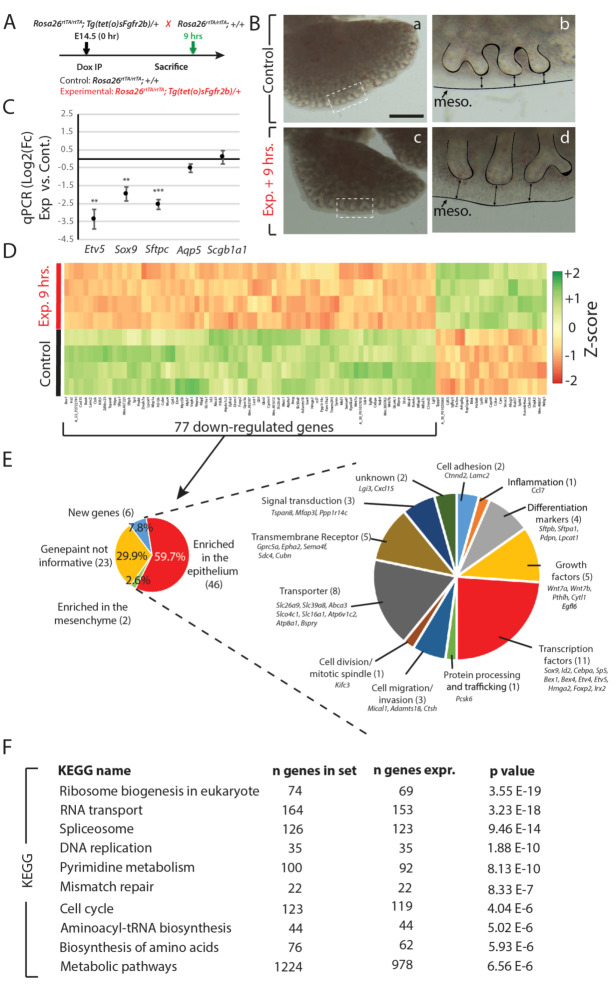
Transcriptomic effects of transient Fgfr2b signaling inhibition for 9 h in E14.5 lungs. (A) Experimental design: pregnant females carrying E14.5 experimental and littermate control embryos were injected with a single Dox-IP and sacrificed 9 h later. (B) Brightfield images of control (a, b) and experimental (c, d) lungs. Note the elongated epithelial tubes, reduced branching, and increased distance between the distal tips and mesothelium (black arrows) in the experimental lung compared to control. (Meso. = mesothelium). Scale bar: (a, c) 500 µm, (b, d) 125 µm. (C) qPCR analysis showing downregulation of Etv5, Sox9, and Sftpc in experimental vs. control lungs, while little change is observed in Aqp5 and Scgb1a1. (n = 3; ** p-value < 0.01, *** p-value < 0.001). (D) Microarray expression data of the top 100 genes (according to the p-value) regulated in experimental vs. control lungs. (E) Gene ontology of the 77 downregulated genes from the heatmap in D. Genes were first grouped based on their region of enrichment in the lung according to expression data from ‘genepaint’. The 46 genes enriched in the epithelium, and thus considered to be direct targets of Fgfr2b signaling, were then grouped according to their primary biological functions. (F) KEGG pathway analyses of the entire microarray data showing the top 10 regulated pathways in the experiment, according to significance.