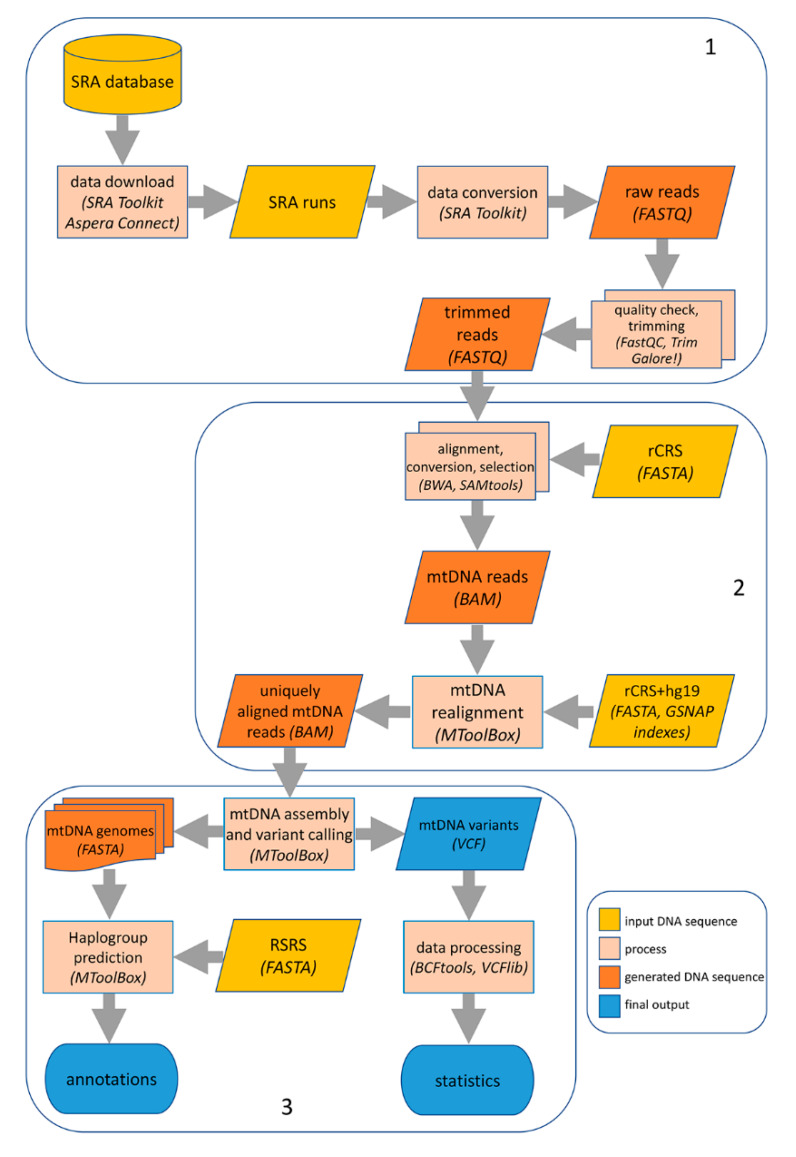
Figure 1.
Computational pipeline for mtDNA analysis in single cell sequencing data. Our computational pipeline comprises three main steps: (1) download from SRA database and preprocessing of raw data from .SRA format to .FASTQ format, followed by quality check and trimming of adapters and low-quality reads; (2) first read alignment onto the rCRS reference sequence and subsequent realignment onto the hg19 (including rCRS) genome to obtain genuine mtDNA reads, using BWA and GSNAP (evoked by MToolBox); (3) mtDNA sequence assembly, variant detection and annotation by MToolBox. Further variant processing was performed with BCFtools and VCFlib.