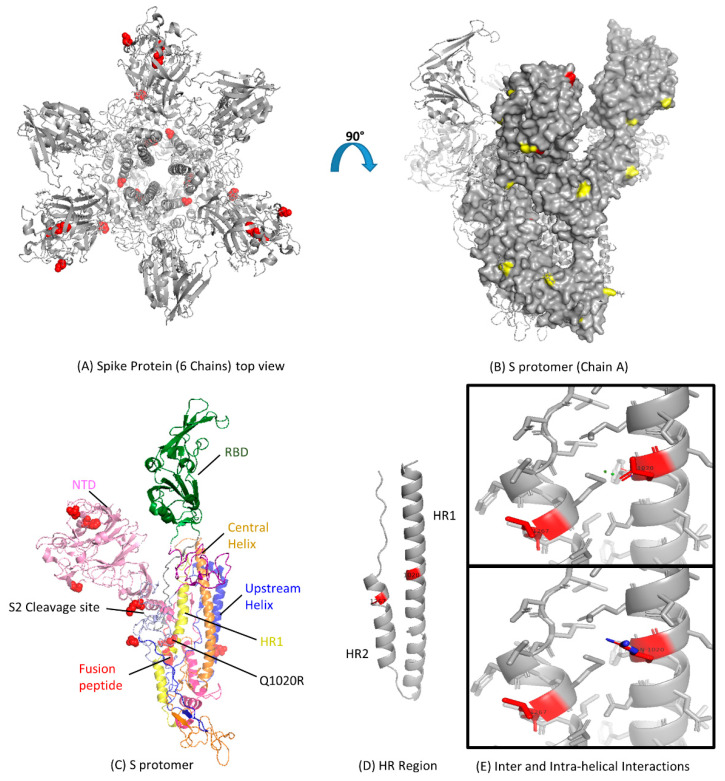
Figure 3.
Functional domains and structural view of amino acid substitutions in the S gene of Middle East Respiratory Syndrome Coronavirus (MERS-CoV). (A) Ribbon representation of the MERS-CoV S gene with amino acid substitutions shown in red spheres with resolved structure in protein database identification (PDB ID: 5 × 59); (B) Surface representation of the S protomer in the Chain A side view showing known glycosylation sites in yellow with amino acid substitutions in red color (PDB ID: 5 × 59); (C) Cartoon representation of all the S protomer for detailed architecture with amino acid changes shown in red spheres; NTD, N-terminal domain; RBD, receptor-binding domain; HR, heptad repeat; CH, central helix (PDB ID: 5 × 59); (D) Ribbon representation of HR region of S protomer; Amino acid changes Q1020R and L1267S are shown in red color (PDB ID: 4MOD); (E) Structural mapping of residue 1020 in the form of inter and intra-helical interactions. The top box shows the interactions for the glutamine residue (Q1020), and the lower box shows the arginine residue (R1020). Hydrogens were removed for a better view. The figures were produced using PyMOL.