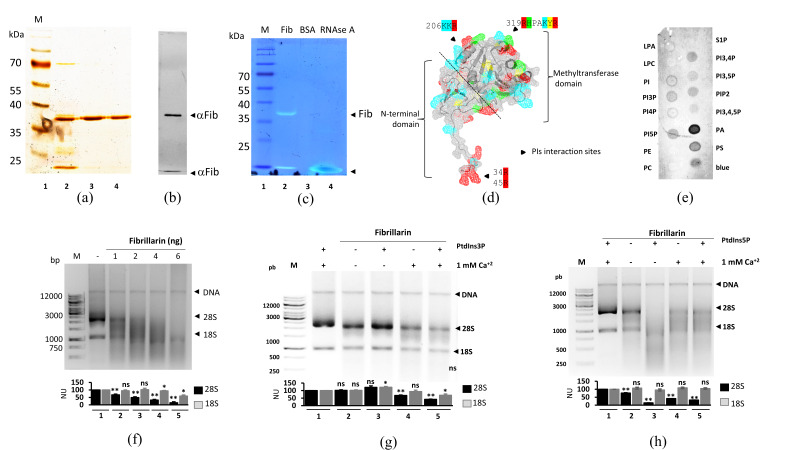
Figure 1.
Fibrillarin acts as a ribonuclease. (a) Silver stain of purified fibrillarin. M: molecular weight marker. 2, 3, and 4: different fibrillarin elution profiles. Only elution 2 and 3 (major quality of protein purification) were used for further analysis. (b) Fibrillarin Western blot revealed two bands marked with arrow-heads that were recognized by specific fibrillarin antibody. (c) In-gel fibrillarin RNA assay shows two signals marked with arrowheads corresponding to fibrillarin (Lane 2). BSA and RNAse A were used as negative and positive controls, respectively (Lanes 3 and 4). (d) Computational inference of possible binding sites for phosphoinositides, one residing in the GAR domain and two in the globular C-terminal domain, are marked with arrows. The fibrillarin sequence with characteristic amino acids for phosphoinositide-binding is noted. Three to five amino acids are necessary for phophoinositide-binding [45,46]. These amino acids should be positively charged with at least one having an aromatic ring. Red: arginine, blue: lysine, green: histidine, yellow: tyrosine. (e) Fibrillarin fat blot assay shows that fibrillarin interacts modestly with several phosphoinositides and strongly with PA. Figure S1 depicts the quantification signals made by ImageJ software. (f) Total RNA in vitro assay was performed while increasing the concentration of protein (from 2 to 8 ng) added to a constant concentration of rRNA (2 μg). Degradation of RNA is directly proportional to the amount of fibrillarin. Copurified DNA from TriZol extraction is indicated by head arrows; the rRNA populations, i.e., 28S and 18S, are indicated. Influence of PI3P (g) and PIP5 (h) on human fibrillarin in vitro ribonuclease assay. (f–h) Quantification of 28S and 18S signals were made by ImageJ software and represented in normalized units (NU), statistical significance was determined by t-test (* p-value < 0.05; ** p-value < 0.01; *** p-value < 0.001, ns= not significant p-value > 0.05) and plots are indicated below the gels from n = 3.