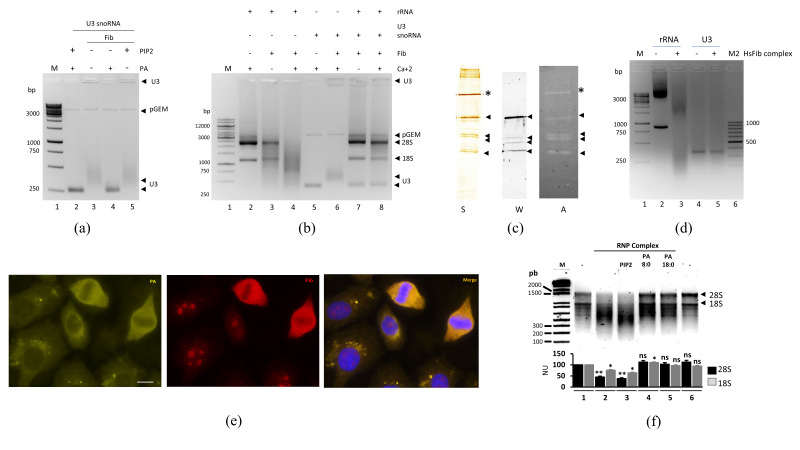
Figure 3.
RNA specific RNA activity by fibrillarin with snoRNA U3. (a) Gel shift of snoRNA U3 with WT fibrillarin. All incubations and procedures carried out at 4 °C, PIP2 and PA were added as indicated. Plasmid control pGEM was used to normalize each lane. (b) RNA activity of fibrillarin with or without guide snoRNA U3 added. Reactions were carried out at 37 °C. (c) In-gel RNA activity of native fibrillarin RNP complex, labeled with silver staining (S), revealed by Western blot (W) and tested by activity assay (A). Arrowheads indicate fibrillarin and potential degradation products, and an asterisk indicates an undetermined ribonuclease activity (d) In vitro RNA activity of HeLa expressed and purified SNAP-tag–fibrillarin complex against total RNA and snoRNA U3. (e) Immunofluorescence showing the colocalization of PA and fibrillarin. The signal corresponds as follows: Blue–DAPI (nucleus signal), Yellow–PA sensor, and RED–fibrillarin. (f) In vitro RNA assay using PA. An inhibition effect was observed using PA containing 8 or 18 carbons and no saturation in the lateral chains (8:0 or 18:0), whereas little effect on the activity was observed with PIP2. Quantification of 28S and 18S signals were made by ImageJ software and statistical significance was determined by t-test (* p-value < 0.05; ** p-value < 0.01; *** p-value < 0.001, ns = not significant p-value > 0.05) and plots are indicated below the gels from n = 3.