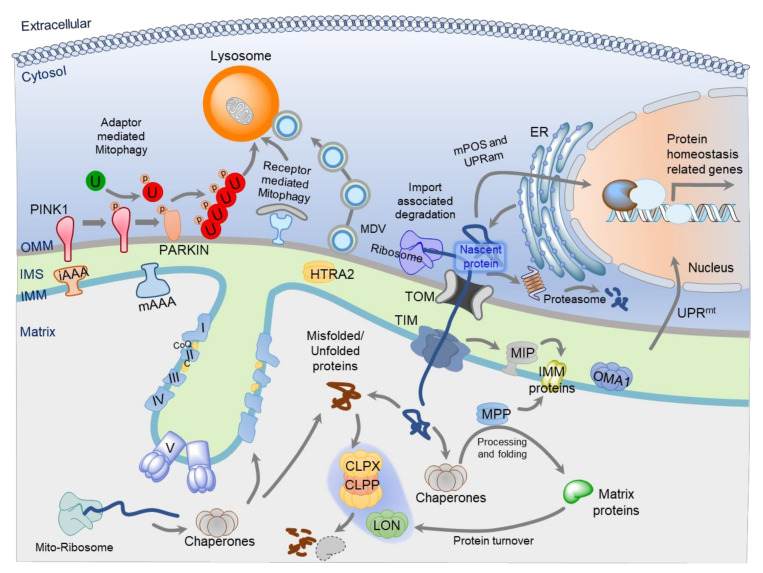
Figure 1.
Overview of the Mitochondrial Protein Quality Control System (MPQC). Mitochondria maintain their protein-quality control at various steps and protect cells against proteotoxic stress. Most of the mitochondrial proteins are synthesized at ribosomes anchored to OMM or ER surface. Nascent mitochondrial proteins are imported into the mitochondria via the TOM/TIM translocase machinery. At the outer face of the OMM, mitochondria have proteostasis system (mPOS, UPRam, and UPS) to ensure that mitochondria maintain their full protein-import capacity. Mitochondrial protein import is assisted by the matrix localized Hsp70 chaperone. Upon import, chaperones together with proteases, ensure the proper trafficking and processing of nascent proteins. Both the nDNA or mtDNA encoded proteins fold into their native conformation, assisted by mitochondrial Hsp70 and Hsp60 chaperone systems. The targeting sequence is cleaved off by the matrix processing peptidase (MPP), giving rise to the mature protein. Some preproteins undergo additional processing by MIP after the removal of their presequence. Under stress or disease conditions, proteins become damaged or denatured. The damaged proteins can be removed by LON and CLPX/CLPP proteases in the matrix; mAAA and iAAA in IMM; and HTRA2 in IMS. Additionally, cells can transcriptionally upregulate the expression of mitochondrial chaperone and protease genes via a mechanism known as UPRmt to relieve stress and re-establish homeostasis. In a failure of these MPQC mechanisms, cells have an alternative mechanism to remove damaged mitochondria. Dysfunctional mitochondria can be removed through the specific autophagy known as mitophagy, and part of mitochondria can be cleared through the mitochondrial-derived vesicles (MDVs). Mitophagy in mammals could be mediated either by a receptor or the PINK1/PARKIN-mediated pathway. Altogether, these mechanisms preserve mitochondrial quality. ER, endoplasmic reticulum; TOM, translocase of the outer membrane; TIM, translocase of the inner membrane; UPS, ubiquitin-proteasome degradation system; mPOS, mitochondrial precursor over-accumulation stress; UPRam, unfolded protein response activated by mistargeting of proteins; MDV, mitochondria-derived vesicles; PINK1, phosphatase and tensin homolog (PTEN)-induced kinase; PARKIN, E3 ubiquitin-protein ligase parkin; U, Ubiquitin; UPRmt, mitochondrial unfolded protein response; iAAA, inner membrane-embedded AAA protease; mAAA, matrix-embedded AAA protease; MIP, mitochondrial intermediate presequence protease; MPP, matrix processing peptidase; OMA1, overlapping activity with m -AAA protease; HTRA2, High-temperature requirement protein A2; CoQ, Coenzyme Q; LON, Lon protease; CLPX, ATP-dependent Clp protease proteolytic subunit X; CLPP, ATP-dependent Clp protease proteolytic subunit P; Mito-Ribosome, Mitochondrial Ribosome.