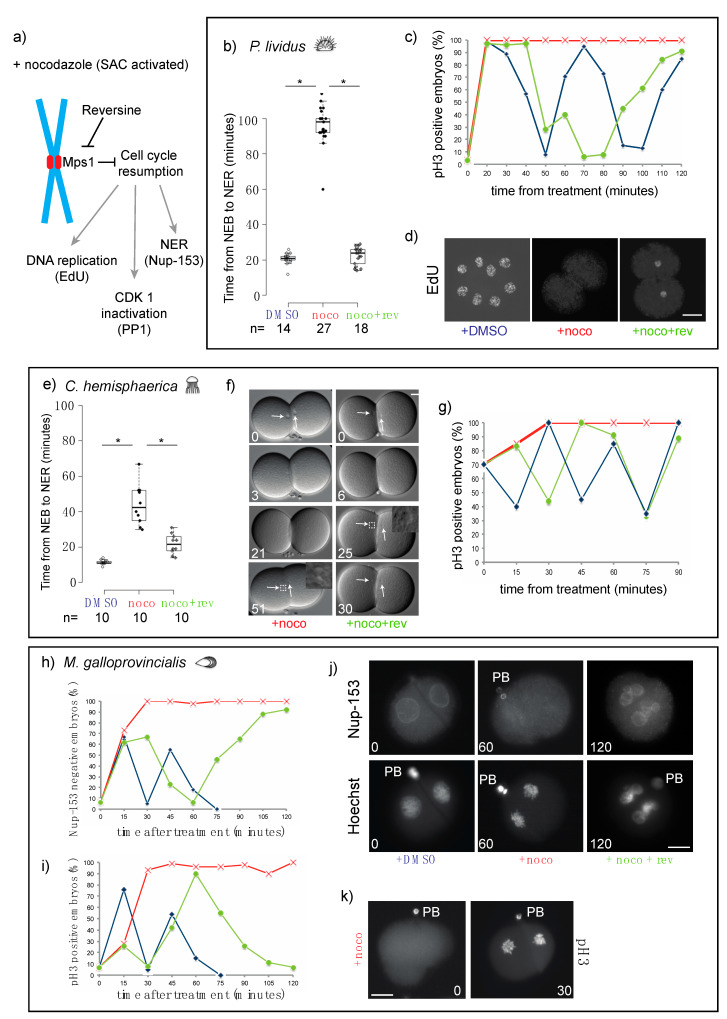
Figure 2.
Microtubule depolymerization causes an Mps1-mediated mitotic block in cleavage stage embryos of P. lividus, C. hemisphaerica, and M. galloprovincialis. (a) Schematic representation of the effect of reversine on cell cycle progression during spindle assembly checkpoint (SAC) activation (+ nocodazole). (b) Quantification of duration of mitosis in P. lividus embryos treated with DMSO, nocodazole, or nocodazole and reversine. Mitosis was measured as time from nuclear envelope breakdown (NEB) to nuclear envelope reformation (NER). Boxes represent 25th–75th percentiles and the median is shown; whiskers mark 5th and 95th percentiles. Each dot represents one cell. Asterisks indicate statistical significance as determined by Student’s t-test, p < 0.001. (c) Quantification of embryos accumulating pH3 in the presence of DMSO (blue), nocodazole (red) or nocodazole/reversine (green) over time equivalent of two cell cycles. (d) Labeling of newly replicated DNA by 5-ethynyl-2′-deoxyuridine (EdU) incorporation in embryos treated with DMSO (left), nocodazole (middle), or nocodazole/reversine (right). (e) Quantification of duration of mitosis in C. hemisphaerica embryos treated with DMSO, nocodazole, or nocodazole/reversine. Box plot parameters as in (b). (f) Representative DIC images of embryos treated with nocodazole (left) or nocodazole/reversine (right). Arrows point at nuclei visible in DIC optics. White squares indicate nuclei-containing regions at time of NER, enlarged in insets. (g) Quantification of pH3 positive C. hemisphaerica embryos in the presence of DMSO (blue), nocodazole (red), or nocodazole/reversine (green). Representative of 4 independent experiments, n = 20–30 for each time point. (h) Quantification of Nup153-labeled and (i) pH3-labeled M. galloprovincialis embryos after treatment with DMSO (blue), nocodazole (red), or nocodazole/reversine (green). (j) Representative images of embryos stained for Nup153 (top), DNA (Hoechst, bottom), and (k) pH3. PB = polar body. Scale bar = 30 µm.