Figure 2 |. DNA repair mechanisms and ovarian cancer.
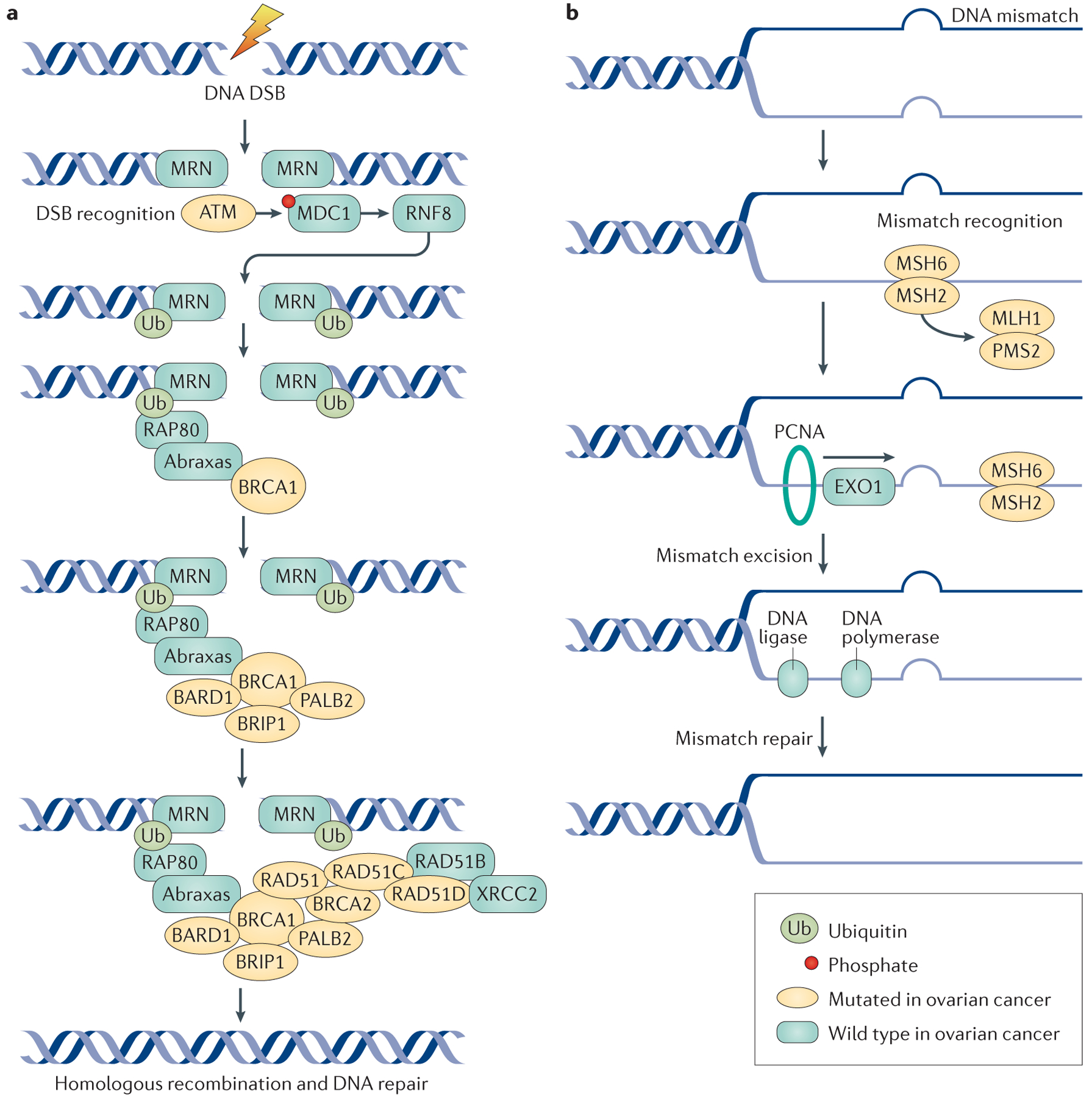
a | The double-stranded DNA break and homologous repair process begins with recognition and sensing of double-strand breaks (DSBs) by the meiotic recombination 11 homologue 1 (MRE11)–RAD50–Nijmegen breakage syndrome protein 1 (NBS1) (MRN) complex, which acts as an activation site for the serine-protein kinase ATM. ATM has a crucial role in DNA repair by coordinating homologous recombination. ATM phosphorylates the histone H2AX, which directly binds to mediator of DNA damage checkpoint protein 1 (MDC1) and NBS1 of the MRN complex, to enhance ATM binding. MDC1 phosphorylation results in a binding site for the E3 ubiquitin-protein ligase RING finger protein 8 (RNF8), which allows ubiquitin-mediated recruitment of downstream DNA damage response proteins, such as receptor-associated protein 80 (RAP80; encoded by UIMC1). RAP80 is an ubiquitin-interaction motif-containing protein that associates with the breast cancer type 1 susceptibility protein (BRCA1) complex through its interaction with Abraxas (encoded by FAM175A); Abraxas is thought to function as a central adaptor protein and contains domains required for BRCA1 interactions. The RAP80–Abraxas complex is crucial for recruiting BRCA1 to the site of DNA repair. BRCA1 and BRCA2 function as scaffolds for other proteins involved in DNA repair. BRCA1-associated RING domain protein 1 (BARD1) and BRCA1-interacting protein 1 (BRIP1; also known as Fanconi anaemia group J protein) bind directly to BRCA1; BARD1 forms a heterodimer with BRCA1, which is essential for mutual stability. BRIP1 also binds to BRCA1 and is required for S phase checkpoint activation. Partner and localizer of BRCA2 (PALB2) helps BRCA1 and BRCA2 bind at sites of DNA damage and helps load RAD51 proteins on to the BRCA proteins; the DNA repair protein XRCC2 is one of the five paralogues of RAD51. Mutations in genes involved in homologous repair lead to defective DNA repair mechanisms, the accumulation of DSBs and an increase in the risk of developing ovarian tumours. b | DNA mismatch repair is mediated by the MutS protein homologue 2 (MSH) proteins, as well as the endonuclease PMS2 and proliferating cell nuclear antigen (PCNA). DNA mismatch repair processes are aberrant in ovarian cancer due to mutations in the genes encoding MutL protein homologue 1 (MLH1), MSH2, MSH6 and PMS2. MSH2 and MSH6 form a heterodimeric complex, which initially identifies mismatched bases and initiates DNA repair. Binding of this complex to the mismatched bases enables the recruitment of MLH1 and PMS2. PCNA attaches to the sites of base mismatch and helps to recruit and tether exonuclease 1 (EXO1; a member of the RAD2 exonuclease family) to the sites of DNA damage. EXO1 excises the mismatched bases, which are then repaired by DNA polymerase and DNA ligase.
