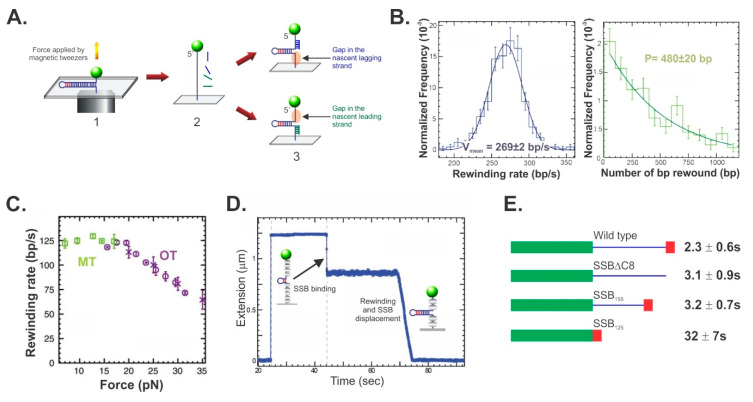
Figure 8.
RecG catalyzes the key steps required for fork regression and displaces SSB in the process. (A) Construction of forks with gaps in either the leading or lagging strands. Here, the 1200 bp hairpin is fully stretched by the application of force from the magnetic tweezers. Then, oligonucleotides complementary to the 5′- or 3′-proximal regions are introduced in separate reactions and allowed to bind. Once the opposing force is decreased, a partial hairpin is extruded and as the oligonucleotide remains annealed to reveal a fork with a gap on the opposite side, either the lagging (top) or leading strand (bottom). (B), RecG rewinds forks with rates and processivity values consistent with a distance-limited fork regression reaction. The reactions rates and processivity of RecG-catalyzed fork rewinding at 37 °C were obtained using the hairpin substrate shown in panel A. P, processivity. (C), RecG can work against large opposing forces during fork rewinding. MT, data from magnetic tweezer experiments. OT, data from assays using optical tweezers; (D), RecG readily displaces SSB during fork rewinding. Here, the hairpin is fully unwound by the application of force. When SSB is added, binding results in the wrapping of the polynucleotide around the tetramers. This causes a shortening of the DNA tether. (E), Functional linkers are required for SSB displacement by RecG during fork rewinding. Each SSB is colored green (N-terminal domain), blue (linker) and red, acidic tip. SSB155 and SSB125 are mutant proteins where the linker was either partially or completely removed, respectively. The values next to each protein, represent the lag time for rewinding to initiate following the addition of RecG. Data panels B-D are from [24] and panel E is from [135].