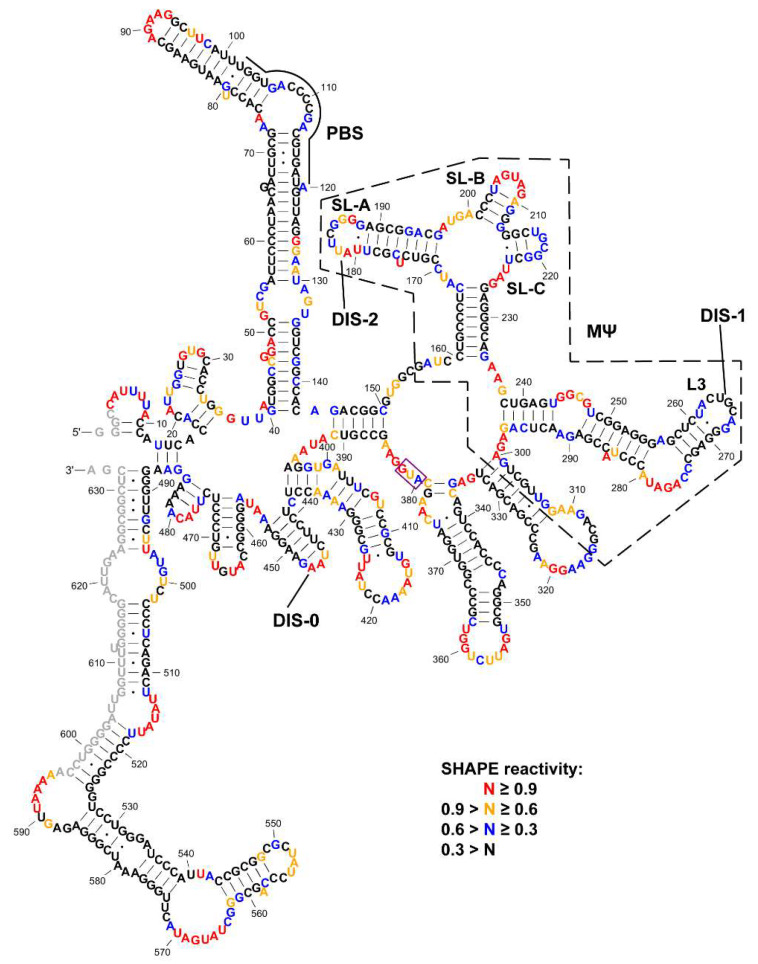
Figure 1.
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) reactivity-constrained lowest energy secondary structure of the 636-nt Rous sarcoma virus (RSV) 5′-leader RNA. The secondary structure model was generated by applying averaged normalized SHAPE reactivity from three independent trials as pseudo free-energy constraints in RNAstructure [53]. Nucleotides are colored in accordance to SHAPE reactivity as indicated in the key. Nucleotides that could not be analyzed due to primer annealing (3′ end) or the lack of resolution (5′ end) are shown in grey. MΨ is indicated by a black dashed box. Stem loops in µΨ and loops that were tested in the dimerization assays (DIS-0, DIS-1/L3, DIS-2/SL-A, SL-B and SL-C) are labeled. The AUG start codon is indicated by a purple box.