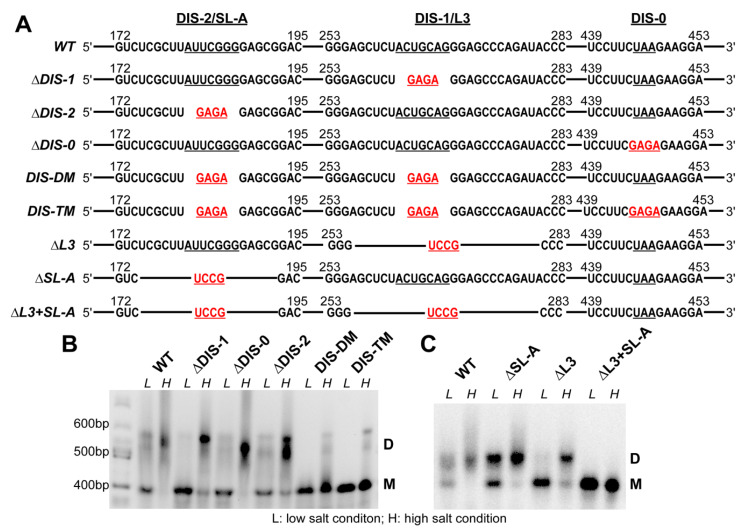
Figure 2.
(A) RNA constructs used in the dimerization studies. Sequences in the regions containing DIS-2/SL-A, DIS-1/L3 and DIS-0 are shown. Nucleotides in the loop regions are underlined. Tetraloop mutations are highlighted in red. (B) Native agarose gel showing the results of dimerization assays of the wild-type (WT) RSV 5′-leader and dimerization initiation signal (DIS) loop mutants. RNAs (0.5 μM) were folded under low salt (L) or under high salt (H), as described in the Materials and Methods. Locations for monomeric (M) and dimeric RNA bands (D) are indicated. This gel shown is representative of three independent replicates. (C) Native agarose gel showing the results of dimerization assays of the WT RSV 5′-leader and more extensive stem loop deletion mutants. The conditions are the same as panel (B). This gel shown is representative of two independent replicates.