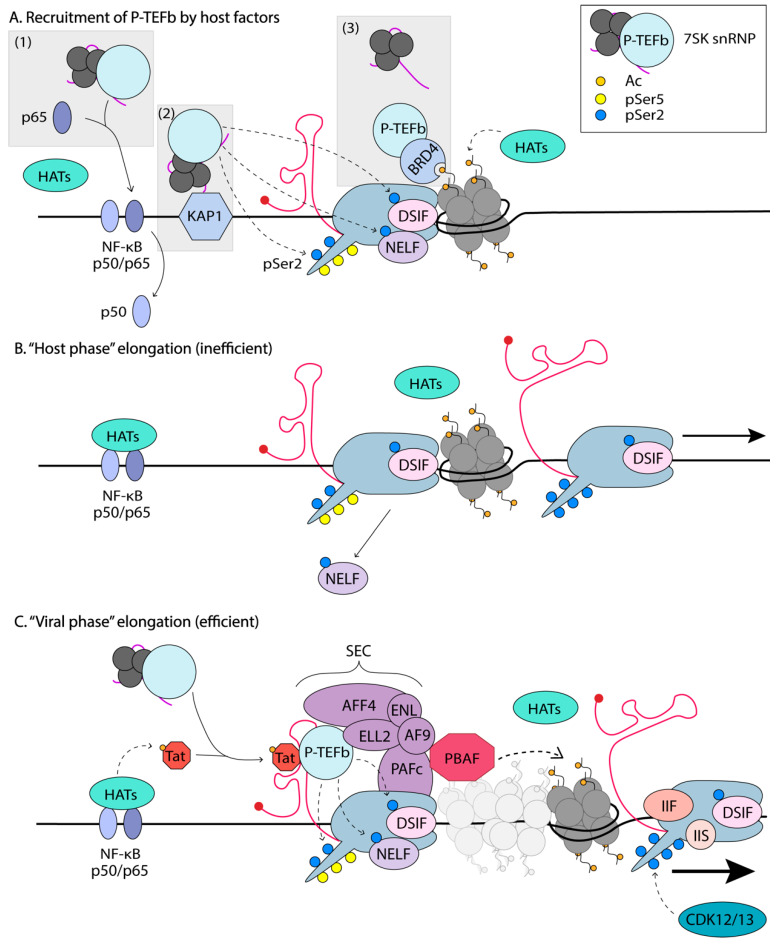
Figure 4.
Regulation of transcription elongation. (A) In activated cells, in the absence of Tat, TFs such as the NF-κB p65 subunit translocate to the nucleus and bind to their consensus sequence. In this process they displace transcriptional repressors, e.g. p50 homodimers and associated HDACs. P-TEFb is recruited (shown by the grey boxes) to the paused RNAPII at promoters, through either a direct interaction with p65 (1), indirectly through recruitment of the 7SK snRNP to promoter DNA by KAP1 (2) or through extraction from the 7SK snRNP by BRD4, which binds to acetylated histones (3). Once in the proximity of the paused RNAPII, the CDK9 subunit of P-TEFb phosphorylates Ser2 residues on the RNAPII CTD, as well as NELF (releasing it from RNAPII) and DSIF (converting it to an elongation factor). (B) In the absence of Tat, the recruitment of P-TEFb is inefficient and results in limited transcription elongation (due to the repressive Nuc-1 positioning), but ultimately leads to a production of some full length HIV-1 mRNAs, which are spliced to produce all transcripts for the proteins that are necessary for the viral lifecycle, including Tat. The level of full-length mRNA production in this “host phase” is dependent on the availability of host TFs, such as NF-κB and NFAT. The efficiency of the “host phase” determines the amount of Tat produced, and whether a certain threshold will be met to enter the “viral phase”. (C) Tat is acetylated by the HAT p300, which enhances the interaction of the CycT1 subunit of P-TEFb with the Tat-TAR recognition motif. Phosphorylation of the CTD, DSIF, and NELF by P-TEFb, triggers the transition of promoter-proximal paused RNAPII to productive elongation. Acetylated Tat also recruits the SEC and the chromatin remodeler PBAF, which further promotes productive elongation through repositioning of Nuc-1. Efficient elongation is maintained through interactions with TFIIS and TFIIF, which relieve polymerase stalling in the gene body. The CTD Ser2 of RNAPII is further phosphorylated, likely by CDK12/13, to promote interactions with RNA splicing and processing factors, leading to efficient gene expression.