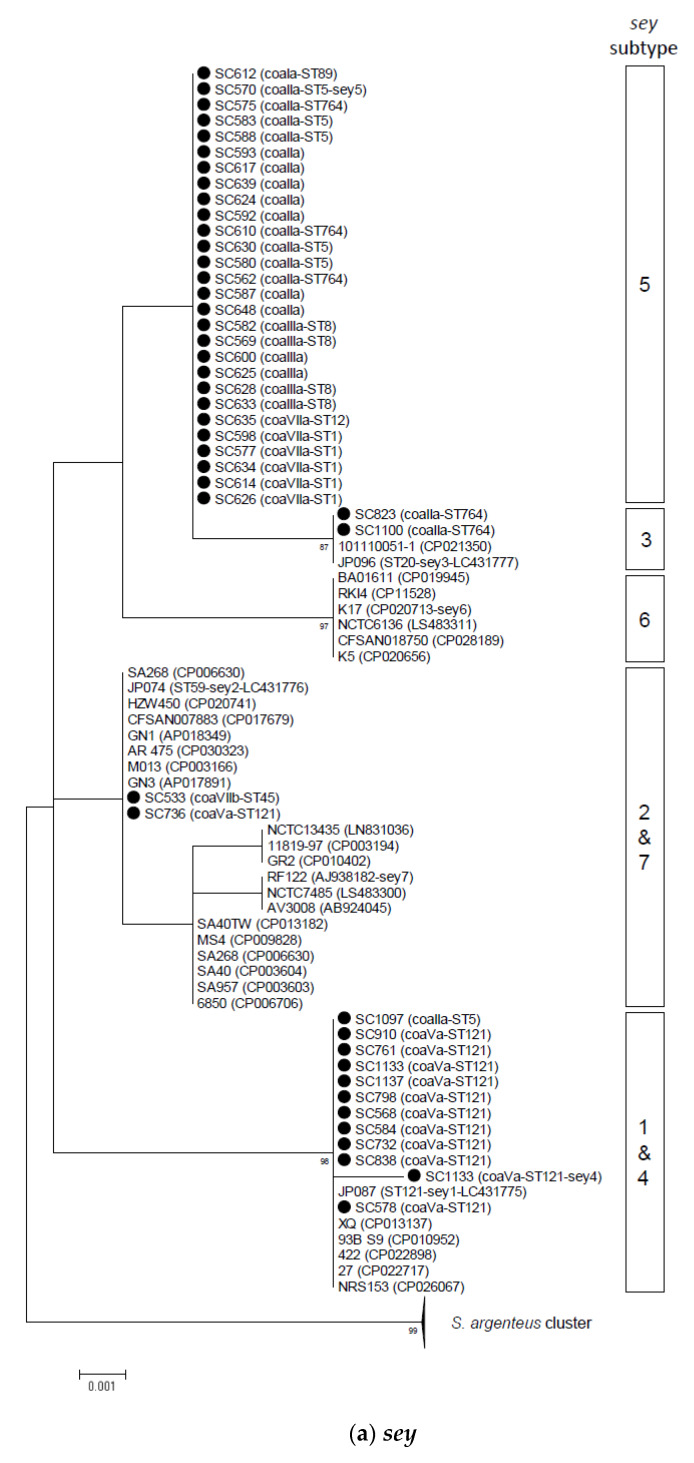
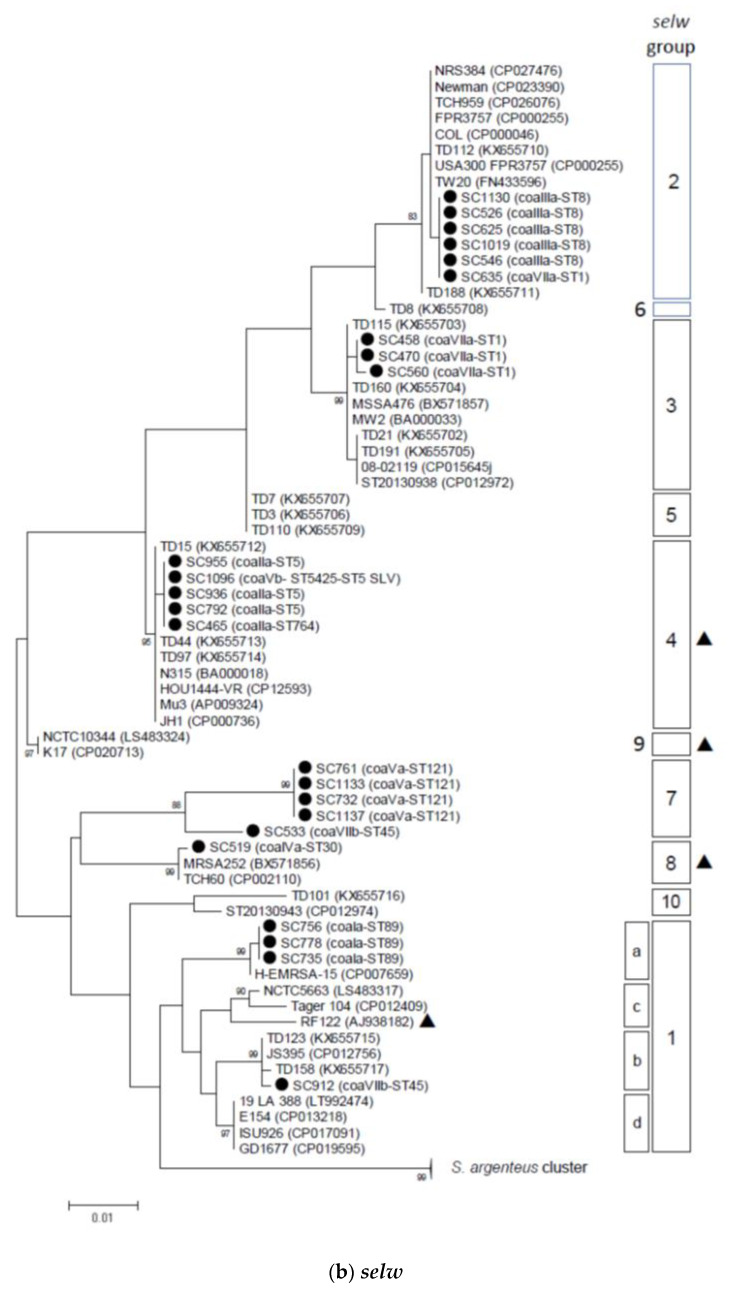
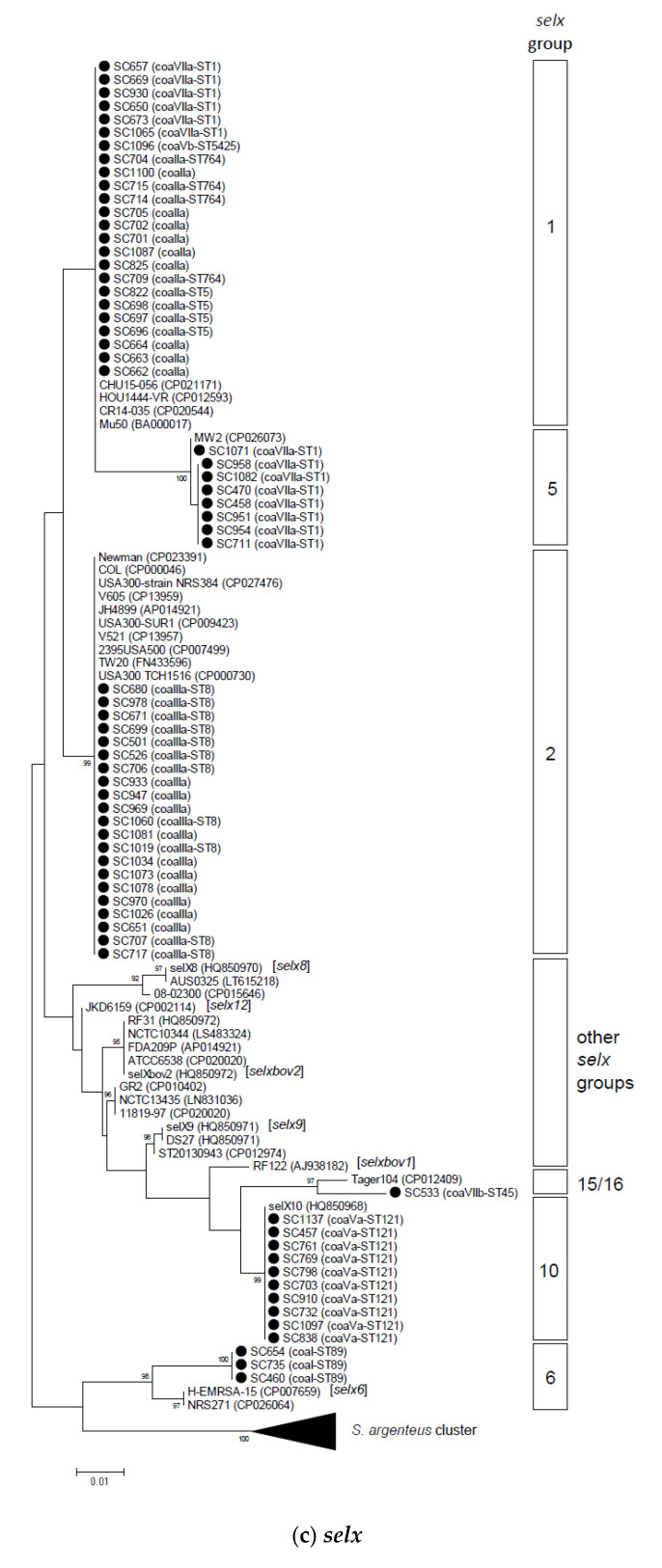
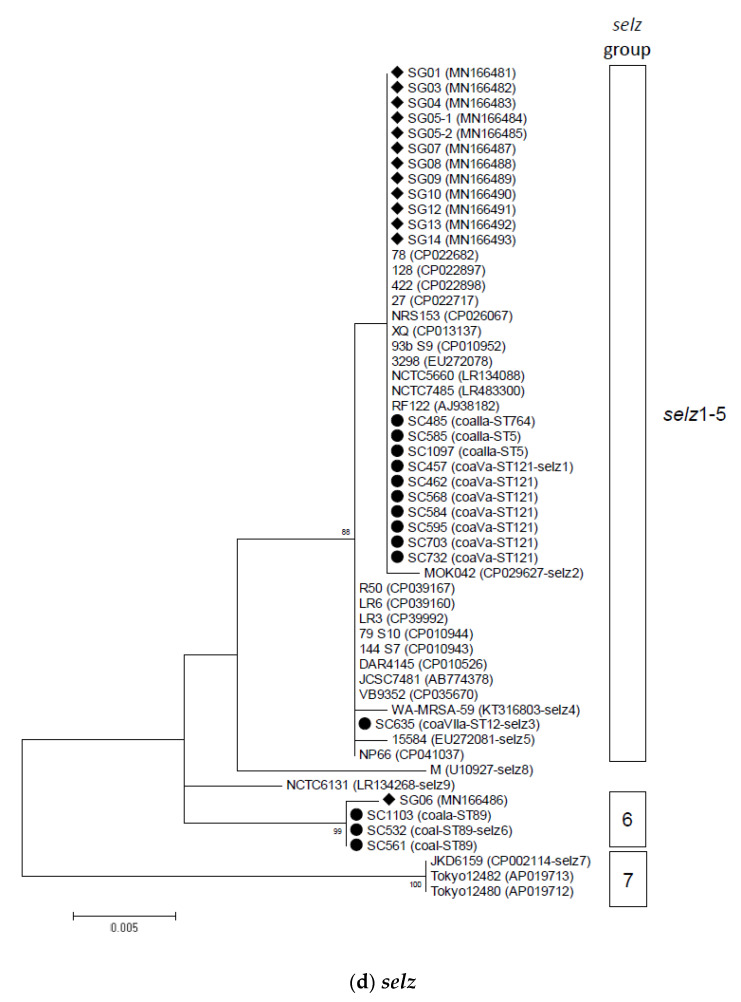
Figure 1.
Phylogenetic dendrogram of sey (a), selw (b), selx (c), selz (d) constructed by the maximum likelihood method using MEGA6. The tree was statistically supported by bootstrapping with 1000 replicates, and genetic distances were calculated by the Kimura two-parameter model. The variation scale is provided at the bottom. The percentage bootstrap support is indicated by the values at each node (values <80 are omitted). The isolates analyzed in the present study are shown with closed circles. The S. argenteus cluster in sey, selw, and selx is shown at the bottom ((a), (b), (c)), while selz of the S. argenteus strain is indicated by a diamond (d). Subtypes/groups of individual SE(-like) genes are shown by boxes on the right. Closed triangles with selw groups and a strain in (b) represent genes encoding truncated proteins. Genotypes of isolates or GenBank accession numbers are shown in parenthesis followed by strain names.