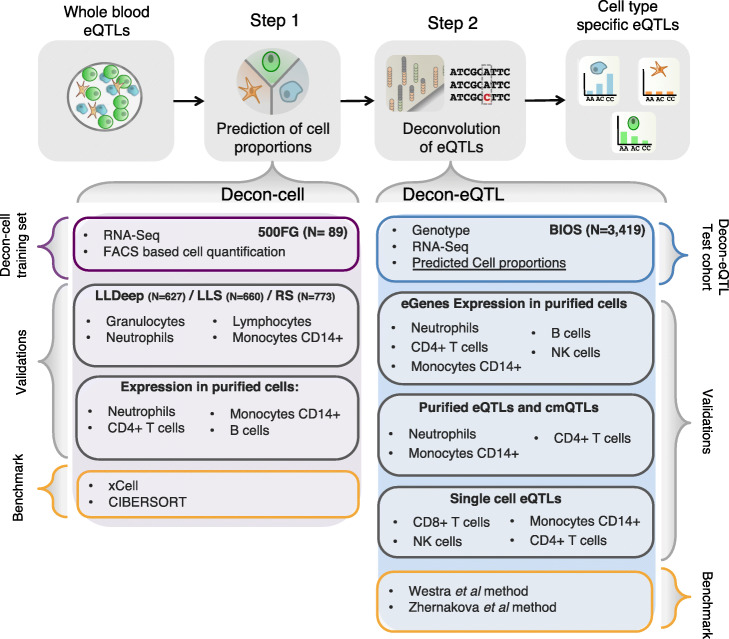
Fig. 1.
Workflow of application of Decon2 to predict cell counts followed by deconvolution of whole blood eQTLs. Using whole blood expression and FACS data of 500FG samples, Decon-cell predicts cell proportions with selected marker genes of circulating immune cell subpopulations. Validations of Decon-cell were carried out on three independent cohorts for which measurements of neutrophils/granulocytes, lymphocytes and monocytes CD14+ were available along with expression profiles of whole blood. Benchmarking of Decon-cell was performed against CIBERSORT [25] and xCell [12]. Decon-cell was applied to an independent cohort (BIOS) to predict cell counts using whole blood RNA-seq. Decon-eQTL subsequently integrates genotype and tissue expression data together with predicted cell proportions for samples in BIOS to detect cell type eQTLs. We validated Decon-eQTL using multiple independent sources, including expression profiles of purified cell subpopulations, eQTLs and chromatin mark QTLs (cmQTLs) from purified neutrophils, monocytes CD14+ and CD4+ T cells [9], and single-cell eQTL results [24]. Benchmarking of Decon-eQTL was carried out for comparison with a previously reported methods that detected cell type–eQTL effects using whole blood expression data, i.e. the Westra et al. [10]