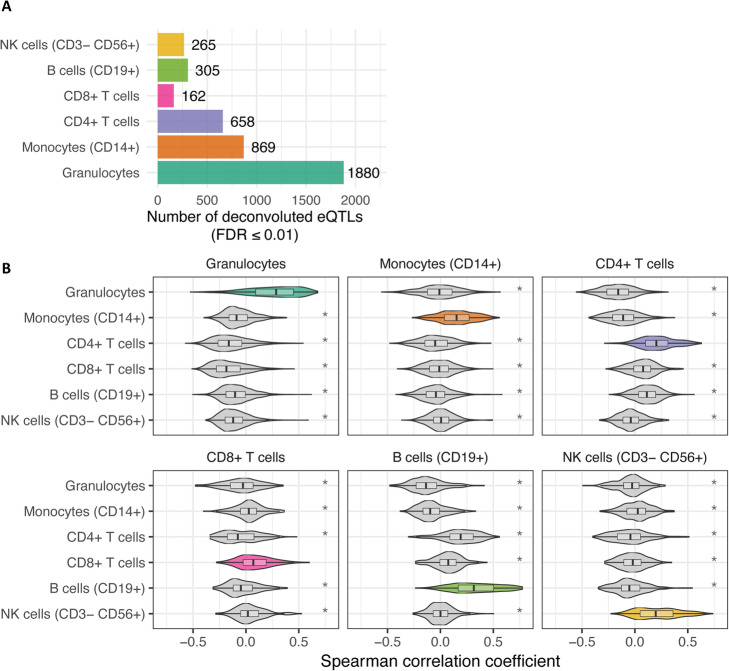
Fig. 3.
Deconvolution of whole blood eQTLs into CTi eQTLs. Decon-eQTL detects CTi eQTLs by integrating proportions of cell subpopulations (predicted by Decon-cell), gene expression and genotype information. a Number of deconvoluted CTi eQTLs in each cell type using whole blood RNA-seq data of 3189 samples in BIOS cohort. b Distribution of Spearman correlation coefficients between expression levels of CTi eQTL genes and cell counts for each cell subpopulation. The CTi eQTL genes show positive and statistically higher correlation (Spearman) with the relevant cell type proportions as compared to the rest (T-test p-value < 0.05) in an independent cohort (500FG)