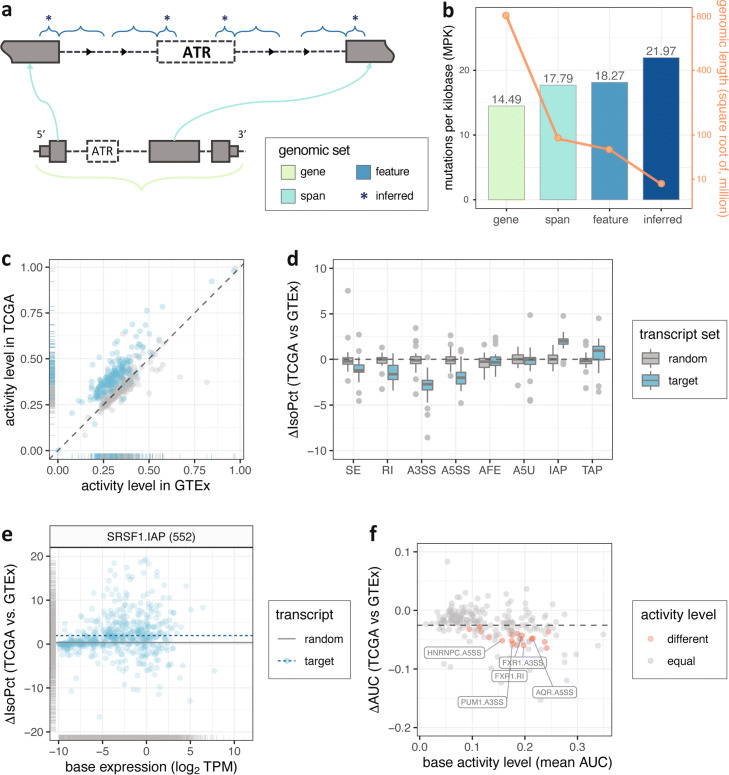
Fig. 6.
Integration of SURF results with large-scale datasets from ICGC, TCGA, and GTEx. a Illustration of RBP location features and three sets of control genomic regions. The set of SURF-inferred features (asterisks) is a subset of all possible location features (blue). The spanning regions (cyan) consist of event body and all possible location features. The set of spanning regions is a subset of the set of all genes (light green). b Mutations per kilobase (MPK) of SURF-inferred features and three control genomic sets. The bars depict the MPK quantified with somatic mutations in ICGC (non-US only) projects. The solid line with the right-hand side axis depicts the total length (in bp) of the genomic regions considered. c Comparison of gene activity level between normal whole blood and LAML tissues. Each dot depicts the mean AUC (activity level) of a gene set in whole blood (GTEx) and LAML (TCGA) samples. The 222 target gene sets are color in blue. The randomized control gene sets are colored in gray. d Changes in isoform percentages across eight ATR event types. Each data point corresponds to either a target transcript set (blue) or a randomized control transcript set (gray). The mean difference in isoform percentages (ΔIsoPct) from normal whole blood (GTEx) to LAML (TCGA) samples is averaged across individual transcript sets. Each of the 222 transcript sets is depicted in one of eight associated ATR event types. e Change in isoform percentages (ΔIsoPct) of 552 SURF-inferred transcript targets of SRSF1 in IAP regulation. A blue dashed line indicates the mean of ΔIsoPct across the full set. The gray solid line indicates the mean of ΔIsoPct across all transcripts in a randomized control set. f Detection of differential activity by discover module of SURF for 222 target transcript sets. Each dot depicts, for one RBP-ATR event type combination, the base activity level (x-axis) and the observed change in the activity level (ΔAUC) from normal (TCGA) to tumor (GTEx) samples. Transcript sets with significant differential activity (FDR <0.05) are marked in red. The gray dashed line indicates the mean of ΔAUC across all control transcript sets