FIGURE 1.
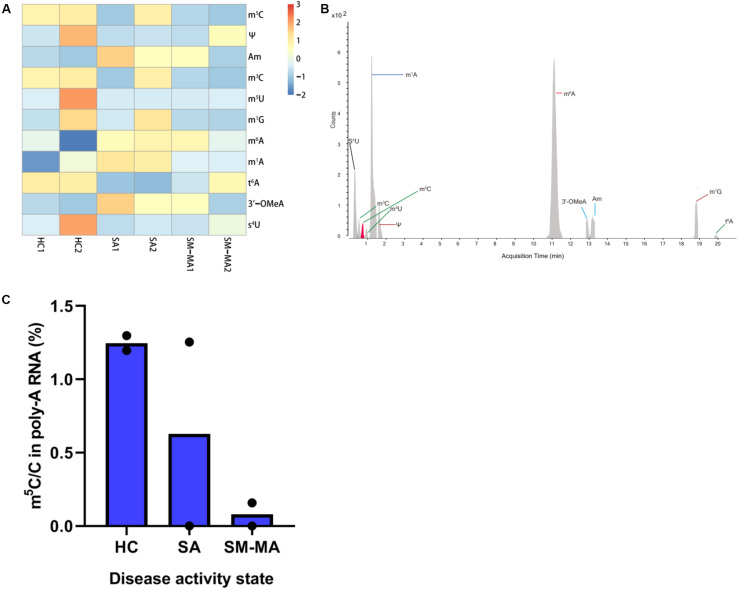
Detection of mRNA modifications by LC-MS/MS among healthy controls (HCs) and systemic lupus erythematosus (SLE) patients with different disease activity. (A) Heatmap of normalized abundance (modification/canonical nucleotide) of 11 mRNA modifications detected by LC-MS/MS between HCs and SLE patients. Red indicates a high z-score, whereas blue indicates a low z-score. (B) LC-MS/MS extracted ion chromatograms of modified nucleotides analyzed in CD4+ T cells mRNA of HCs and SLE patients. S4U, 4-thiouridine; m3C, 3-methylcytosine; m5C, 5-methylcytosine; m5U, 5-methyluridine; m1A, N1-methyladenosine; Ψ, pseudouridine; m6A, N6-methyladenosine; 3′-OMeA, 3’-O-methyladenosine; Am, 2’-O-methyladenosine; m1G, 1-methylguanosine; t6A, N6-threonylcarbamoyladenosine. (C) Global dynamics of calibrated m5C/C levels in HCs and SLE patients with different disease activity (SA, SM-MA). Bars show the mean of individual biological replicates (n = 2).