Abstract
Blocking the interaction between phosphotyrosine (pTyr)-containing activated receptors and the Src homology 2 (SH2) domain of the growth factor receptor-bound protein 2 (Grb 2) is considered to be an effective and non-cytotoxic strategy to develop new anti-proliferate agents due to its potential to shut down the Ras activation pathway. In this study, a series of phosphotyrosine containing cyclic pentapeptides were designed and synthesized based upon the phage library derived cyclopeptide, G1TE. A comprehensive SAR study was also carried out to develop potent Grb2-SH2 domain antagonists based upon this novel template. With both the peptidomimetic optimization of the amino acid side-chains and the constraint of the backbone conformation guided by molecular modeling, we developed several potent antagonists with low micromolar range binding affinity, such as cyclic peptide 15 with an Kd = 0.359 μM, which is providing a novel template for the development of Grb2-SH2 domain antagonists as potential therapeutics for certain cancers.
Keywords: Grb2-SH2 domain, Cyclic peptides, G1TE analogs, Phosphotyrosine, Grb2-SH2 antagonist
Growth factor receptor-bound protein 2 (Grb2) is an essential adapter protein, possessing a single Src homology 2 (SH2) domain flanked by two SH3 domains.1 Grb2-SH2 domain directly mediates erbB-2 (HER-2/neu) signaling by binding to pTyr motifs and the downstream activation of mitogenic Ras pathways, which have been implicated in the etiology of certain breast cancer.2,3 Grb2-SH2 domain is an ideal target for drug development,4,5 in that the phosphopeptide ligands that bind it are required to adopt a β-turn conformation, which has been demonstrated by both NMR and X-ray analysis.6–8 Molecules that can selectively antagonize the Grb2-SH2 should interrupt these signaling pathways. These agents may act as therapeutic leads for diseases, such as cancer, in which the Ras signaling pathway plays a major role.9–12
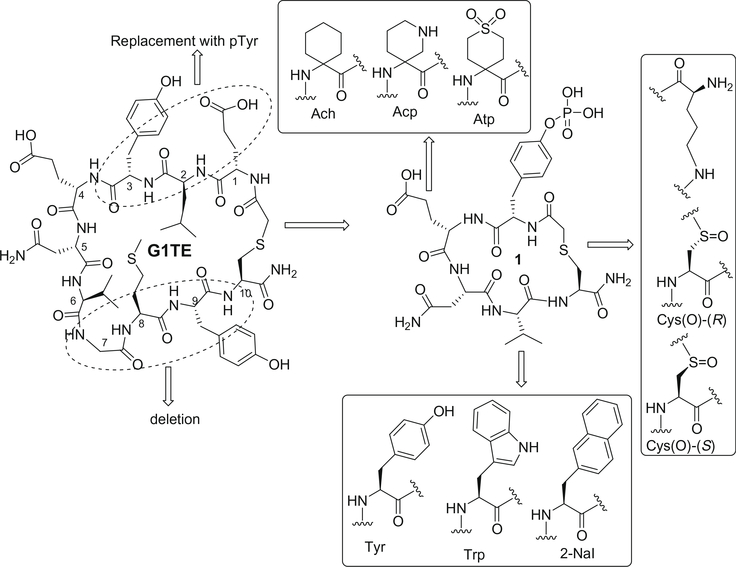
In our previous studies, we discovered a novel disulfide-bridged cyclic peptide G1, generated by phage library.13 This peptide was comprised of the sequence -Glu-Leu-Tyr-Glu-Asn-Val-Gly-Met-Tyr-, bracketed by two terminal cysteines that formed the cyclic peptide through a disulfide linkage. Subsequently, we further designed and synthesized its redox stable thioether-bridged analogue, cyclo (CH2CO-Glu1-Leu2-Tyr3-Glu4-Asn5-Val6-Gly7-Met8-Tyr9-Cys10)-amide, termed G1TE (Fig. 1), which has shown comparable binding affinities (IC50 = 15–20 μM) to the Grb2-SH2 domain protein, in Biacore binding assays.13 Based on the G1TE, a number of redox-stable thioether-cyclized analogs were developed, some of which exhibited potent binding affinity.14–19 G1TE and its analogues may form a largest circle-like binding surface when they bind to Grb2-SH2 domain, as suggested by molecular modeling and NMR studies.20,21 Molecular modeling of the complex structure of Grb2-SH2 protein with G1TE indicated that the side chains of the amino acid residues in the 1, 3 and 5 positions of G1TE form strong hydrogen-bond interactions with the Arg 67 (αA-helix) and Arg 86 (βC-strand) residues and the three Ser residues 88, 90, and 96, and Lys 109 and Leu 120 within the binding pocket. However, the amino acid residues in the 2, 7, 8, and 9 positions of G1TE do not interact with the Grb2-SH2 protein. To further improve the affinity and reduce the peptide character of the novel Grb2-SH2 domain antagonists, we were interested in developing a new scaffold with smaller ring size but higher affinity. We assumed that keeping essential amino acids and deleting unnecessary amino acids of G1TE would increase the binding affinity and selectivity. Replacement of Glu1, Leu2 and Tyr3 of G1TE with p-Tyr would increase hydrogen-bond interaction with the Arg 67 and Arg 86 of the SH2 domain binding cavity; on the other hand, the phosphoryl group itself would undergo additional intramolecular hydrogen-bond interactions, which stabilizes the binding conformation of cyclic peptide. This is a brand new strategy distinct from any approaches reported previously to improve the selectivity and affinity. This article reports our recent discovery of potent tyrosine-phosphorylated G1TE analogues by overall side-chain and backbone modifications and conformational constraint (Fig. 1).
Figure 1.
Molecular design of Grb2-SH2 domain antagonists based upon thioether-bridged cyclic peptide G1TE.
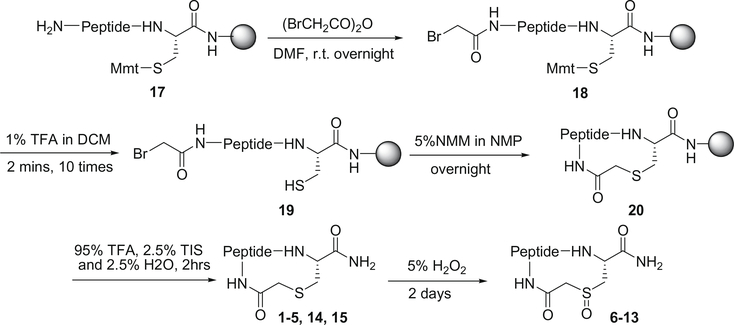
The cyclic peptides 1–15 in Table 1 were synthesized by using the procedure described in Fig. 2. The linear peptide precursors were synthesized on an ABI 433A peptide synthesizer, starting with Rink Amide resin for establishing the C-terminal carboxamide, and using the chemical protocols based on the Fmoc chemistry. The incorporation of N-Fmoc-Tyr[PO(OH, OBzl)-OH was carried out manually and coupled twice using HATU as the coupling reagent. After completion of peptide elongation, the Mmt protecting group on Cys was removed by treating with 1% TFA in DCM to afford the resin bound peptide 19. Subsequently, cyclic peptides were formed by using NMM as a base. The resulting cyclopeptide bound resins were washed six times with NMP, DCM, and MeOH, and then dried under reduced pressure overnight. The cyclopeptides were cleaved from the resin and fully deprotected by treating with 2.5% TIS and 2.5% H2O in TFA (10 mL/g resin) for 2 h. The oxidation of the thioether functionality into sulfoxide was accomplished using a 5% H2O2 aqueous solution. The resulting two isomers were easily separated by RP-HPLC. All peptides were purified to homogeneity by preparative RP-HPLC (Vydac C18 column), and their structures were confirmed by MALDITOF-MS mass spectrometry. The absolute configuration of the sulfoxide diastereoisomers was determined by CD spectra using documented method.22–26
Table 1.
Grb2-SH2 domain inhibitory activity of the G1TE analogs 1–16 with the general structure cyclo-(pTyr1–AA2–Asn3–AA4–AA5)-amidea
| Compds | Modifications of amino acid residues |
Activity Kdb (μM) | ||
|---|---|---|---|---|
| AA2 | AA4 | AA5 | ||
| G1TE | – | – | – | 25 |
| 1 | Glu | Val | Cys-CH2CO- | 3.6 (±0.0.01) |
| 2 | Ach | Val | Cys-CH2CO- | 1.42 (±0.04) |
| 3 | Ach | Tyr | Cys-CH2CO- | 1.06 (±0.03) |
| 4 | Ach | Trp | Cys-CH2CO- | 1.05 (±0.03) |
| 5 | Ach | 2-NaI | Cys-CH2CO- | 2.71 (±0.09) |
| 6 | Ach | Val | Cys(O)-(R)-CH3CO- | 1.56 (±0.05) |
| 7 | Ach | Val | Cys(O)-(S)-CH2CO- | 1.76 (±0.04) |
| 8 | Ach | Tyr | Cys(O)-(R)-CH3CO- | 1.36 (±0.04) |
| 9 | Ach | Tyr | Cys(O)-(S)-CH2CO- | 1.72 (±0.05) |
| 10 | Ach | Trp | Cys(O)-(R)-CH3CO- | 2.14 (±0.06) |
| 11 | Ach | Trp | Cys(O)-(S)-CH2CO- | 1.29 (±0.03) |
| 12 | Ach | 2-Nal | Cys(O)-(R)-CH3CO- | 4.8 (±0.0.03) |
| 13 | Ach | 2-Nal | Cys(O)-(S)-CH2CO- | ndc |
| 14 | Atp | Val | Cys-CH2CO- | 0.88 (±0.20) |
| 15 | Acp | Val | Cys-CH2CO- | 0.359 (±0.06) |
| 16 | Ach | Val | Lys | 7.55 |
The binding affinity of these compounds was evaluated by Biacore assays.
Values are means of two experiments, standard deviation is given in parentheses.
nd = not determined.
Figure 2.
General procedure for the synthesis of thioether and sulfoxide bridged cyclic pentapeptides.
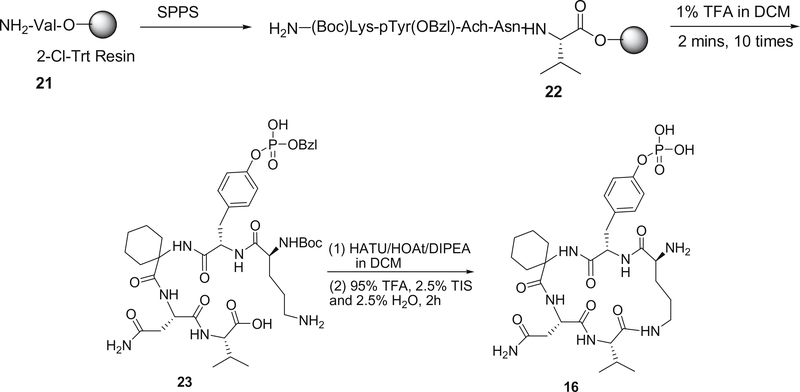
The synthesis of cyclic peptide 16 was described in Fig 3. The linear peptide precursors H-(Boc)Lys-pTyr(OBzl)-Ach-Asn-Val-2-Cl-Trt resin were synthesized by Fmoc SPPS. After the peptides were cleaved from resin, the head-to-tail macrocyclization of the linear peptide was achieved successfully using HATU as the coupling reagent and HOAt as the accelerator27 and then deprotected with the TFA–TIS–H2O cocktail. After side-chain deprotection with the TFA–TIS–H2O cocktail, the cyclic peptide 16 was obtained in 30% overall yield.
Figure 3.
Synthetic route of cyclic pentapeptide 16.
Docking studies of these 16 compounds were performed on the crystal structure of Grb2-SH2 domain (PDBID: 1TZE),6 to which the KPFpYVNV peptide bound, using the program Glide 4.5,28–31 part of the First Discovery suite (Maestro). The starting structure for the protein was prepared and fixed by the Protein Preparation Wizard, a comprehensive protein preparation facility. All water molecules were deleted; the bound ligand (KPFpYVNV) was adjusted manually; bond orders were assigned, and then hydrogen was added to all atoms in the structure. The orientation of hydroxyl groups, amide groups of Asn and Gln, and charge state of His residues were optimized using the utility protassign. The final step was a restrained minimization by the impref utility until the average root-mean-square-deviation (rmsd) of the hydrogen atoms reached 0.3 Å, leaving heavy atoms in place. In order to study the binding modes of the antagonists, grids files representing shape and properties of the receptor were generated. The three-dimensional structures of the compounds 1–16 were constructed using the builder tool available through Maestro interface by modifying the known peptide bound in 1TZE. The initial geometries of the structures were optimized using the OPLS_2005 force field, performing 5000 steps of conjugate gradient minimization. The compounds were subjected to flexible docking using the pre-computed grid files. Glide 4.5 has three docking precision mode: high-throughput virtual screening (HTVS), standard precision (SP) and extra precision (XP). Here, to try to get the closet binding conformations, we used XP mode.
To try to explain the biological activity differences between cyclic peptides, conformational searches were adopted. The software used was MACROMODEL 9.5,32 and the method utilized was Torsional sampling (Monte Carlo Multiple Minimum, MCMM), with the OPLS_2005 force field. Most experimental studies are carried out in solvent, and the solvent medium can have a major effect on molecular structures and energies, especially for molecules having several polar functional groups, like those we study here. So when doing conformation searching, we used a called GB/SA model to treat the solvent as a fully equilibrated analytical continuum starting the near the van der Walls surface of the solute. The parameters were set according to the following: the energy window for saving structures was 50 kJ/mol; the maximum iterations for minimizing the molecules were 5000; the convergence threshold was 0.05; and the others adopted the default values.
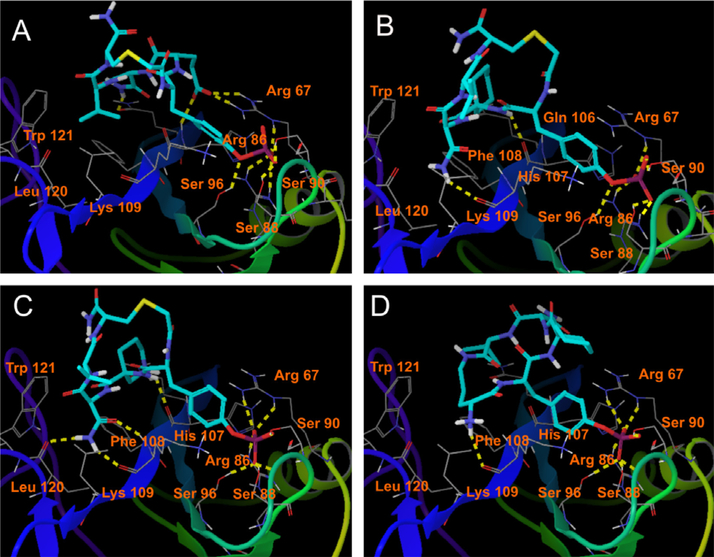
It has been well documented that phosphotyrosine is important for linear peptide ligand binding to Grb2-SH2 domain by forming extensive hydrogen bonds and hydrophobic interactions.6,33 Deleting unnecessary amino acids of G1TE, and phosphorylation of Tyr3 of G1TE provided for us peptide 1 that competitively bound to the Grb2-SH2 domain protein with an Kd of 3.6 μM. The inhibitory activity of pTyr-containing cyclic pentapeptide 1 is at least 7 times more potent than the corresponding G1TE. Molecular modeling indicates that the negatively charged pTyr oxygens of cyclic pentapeptide 1 form key binding interactions with the Arg 67 and Arg 86 and the three Ser residues 88, 90 and 96 of the protein, and consensus amino acid Glu 2 of the cyclic pentapeptide 1 form two extra hydrogen bonds with residue Arg 67 because of the strong positive-negative charges attraction (Fig. 4A).
Figure 4.
Docking poses of cyclic peptides 1 (A), 2 (B), 15 (C, R enantiomer) and 16 (D) with Grb2-SH2 domain (PDB ID 1TZE). Hydrogen bonds between ligands and protein are represented by yellow dot lines. For clarity, only polar hydrogen atoms are shown here.
The X-ray structure of Grb2-SH2 domain complexed with pTyr-containing peptide ligand has shown that the phosphopeptide binds to Grb2-SH2 domain in a β-turn conformation due to the existence of Trp121 around the asparagine binding pocket.6 Therefore, we intentionally introduced a turn-inducing amino acid, such as 1-amino-1-cyclohexylic acid (Ach), in the position 2. When replacing Glu 4 with Ach 4, the resulting peptide 2 loses charge interactions with Arg 67, but gains hydrophobic Van der Waals interactions with Gln 106 and Phe 108 of the Grb2-SH2 domain, as shown in Figure 4B. In addition, the 310 helix turn, with the torsion angles φ = −62.1°, ψ = 34.3° and ω = 170.4°, induced by Ach is also favored for the peptide binding to the Grb2-SH2 domain. The overall effect makes peptide 2 (Kd = 1.42 μM) approximately 2.5-fold more potent than peptide 1.
By analyzing the model of the complex structure of G1TE with Grb2-SH2 domain, we found that Val 4 has van der Waals interactions with the side chains of Lys 109 and Leu 120 of the Grb2-SH2 domain. In addition, Val 4 localizes in a hydrophobic region of the Grb2 protein, which is composed of Phe 108, Gln 106, Trp 121, Lys 109, and Leu 120. Therefore, introducing a more hydrophobic amino acid residue into position 4 of cyclic pentapeptide 2 would improve this kind of van der Waals interactions. Considering these characteristic interactions, we incorporated a series of hydrophobic amino acids in position 4, as shown in Table 1. The binding affinity of the modified peptides 3 (Kd = 1.06 μM) and 4 (Kd = 1.05 μM) increases approximately 1.5-fold when replacing Val 4 with tyrosine (Tyr) or tryptophan (Trp). Compared to valine, these amino acids are more hydrophobic, which results in favorable and well defined Van der Waals interactions. However, these improved interactions can be overweighed by increasing exposed hydrophobic surface area under some circumstances. For instance, the binding affinity of the resulting peptides 5 (Kd = 2.71 μM) was not improved, and even decreased when Val 4 was substituted with L-2-naphthylalanine (2-Nal).
To further constrain the backbone conformation of cyclic pentapeptides, we also tried to modify the thioether bridge of these peptides, since this –CH2–S–CH2– motif is conformational flexible and may contribute to an entropic penalty for the thioether-bridged cyclic pentapeptides binding to Grb2 protein. When the thioether-cyclized peptide 2 was oxidized into the relatively more rigid R-configured sulfoxide 6 and S-configured sulfoxide 7, the binding affinity was slightly decreased (6, Kd = 1.56 μM; 7, Kd = 1.76 μM) than its precursor thioether analog 2, probably due to the disfavored change of the peptide backbone conformation. This trend was also observed in the Tyr 4, Trp 4, and Nal 4-containing peptides 3, 4, 5, 8, 9, 10, 11 and 12. Compared to thioether-bridged analogues, the activities of these sulfoxide bridged cyclopeptides are not improved, due to the steric hindrance caused by the bulky side chain of tyrosine, tryptophan, and 2-naphthylalanine, respectively. We also incorporated Polyamide linkage into cyclic pentapeptides 2, and examined the potency to determine the optimal linkage which favors forming the β-turn like conformation. The binding affinity of the resulting peptide 16 (Kd = 7.55 μM) was 5.3 times less than its precursor thioether analog 2. This indicates that the rigid cyclization constraint conferred by the backbone cyclization disfavors either the formation of β-turn conformation or the optimal orientation of the side chains of essential residues. These results indicate that the linkage in cyclic pentapeptides, which are not involved in the direct interactions with the binding pocket of the protein, still has a function to induce the required conformation via its backbone.
4-Amino-1,1-dioxo-hexahydro-thiopyran-4-carboxylic acid (Atp) and 3-amino-piperidine-4-carboxylic acid (Acp) are turn-inducing amino acids, similar to Ach. We replaced Ach 2 with Atp and Acp, respectively, whereby the resulting cyclic peptides 14 and 15 showed 1.6 and 4 times more inhibitory activity than the corresponding unmodified peptide 2, respectively (14, Kd = 0.88 μM; 15, Kd = 0.359 μM). Molecular modeling shows that that the replacing would induce more hydrogen-bond interactions with Leu 120and Lys 109 of the Grb2-SH2 domain (Fig. 4C).
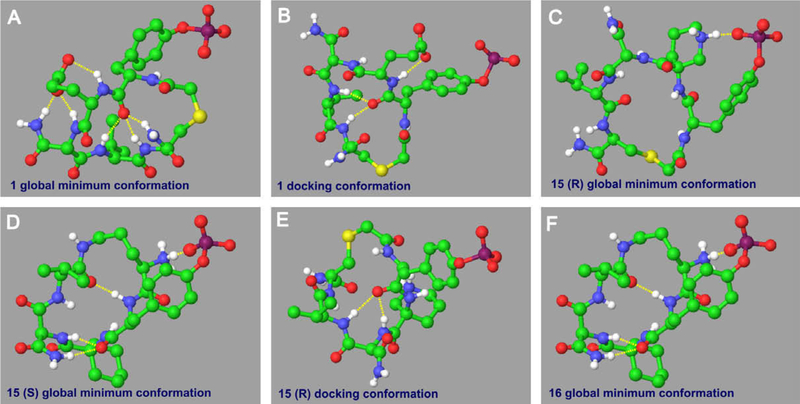
There are two intramolecular hydrogen bonds, which are formed by a same oxygen atom with two same hydrogen atoms in the 18 member ring, in almost all these 16 compounds’ docking conformations (Fig. 5E is as an example), except that cyclic peptide 1 has three (Fig. 5B). This extra hydrogen bond is created by the propanoic acid group with the amide group sitting next to it. But for the global minimum conformations we searched, the situations are different. Cyclic peptide 15, actually, is a racemic mixture. For its R enantiomer, the global minimum conformation has only one intramolecular hydrogen bond, produced by the phosphoric acid group and the charged amino atom in the rigid six-member ring (Fig. 5C). Whereas, its S enantiomer’s global minimum conformation has four hydrogen bonds (Fig.5D), which makes it much more difficult to change its global minimum conformation to binding conformation than R enantiomer. So we can conclude that R enantiomer of cyclic peptide 15 has stronger binding affinity than S enantiomer just from conformational point. Cyclic peptide 1 can even produce six intramolecular hydrogen bonds (Fig. 5A), among which three are through the flexible propanoic acid group; Cyclic peptide 16 can produce four (Fig. 5F), one of which is formed by the negative charged phosphoric acid group with positive charged amino group and therefore is much stronger than other hydrogen bonds. So, for the same reason, for cyclic peptides 1 and 16, it is also much harder to change their global minimum conformations to binding conformations than other analogues. This, from the other side, explains why cyclic peptide 15 has best and cyclic peptides 1 and 16 have worst biological activities toward Grb2-SH2 domain.
Figure 5.
Conformation analyses of compounds 1, 15 and 16. Intramolecular hydrogen bonds are represented by yellow dot lines. For clarity, only polar hydrogen atoms are shown here.
In summary, we have designed and synthesized a series of phosphotyrosine containing cyclic pentapeptides through deleting unnecessary amino acids, based upon molecular modeling of the complex structure of Grb2-SH2 protein with G1TE. By introducing a turn-inducing Acp in 2th position, a hydrophobic and less flexible valine in the 4th position, and keeping thioether linkage respectively, we developed a globally and locally optimized inhibitor 15 with a Kd = 0.359 μM, as evaluated by Biacore assays. This fully elaborated small peptidomimetic provides us a novel template for the development of pharmacokinetically improved peptidomimetics that can selectively bind to Grb2-SH2 domain, and may act as potential therapeutic agents for the treatment of erbB2-related cancers.
Supplementary Material
Acknowledgments
This project was supported by the National Natural Science Foundation (Grant no. 20802078), National Basic Research Program of China (973 Program, Grant no. 2009CB940900), the Guangdong Natural Science Foundation (Grant no. 8151066302000008) and the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bmcl.2009.03.134.
References and notes
- 1.Margolis B Prog. Biophys. Mol. Biol 1994, 62, 223. [DOI] [PubMed] [Google Scholar]
- 2.Yip SS; Crew AJ; Gee JM; Hui R; Blamey RW; Robertson JF; Nicholson RI; Sutherland RL; Daly RJ Int. J. Cancer 2000, 88, 363. [PubMed] [Google Scholar]
- 3.Lim SJ; Lopez-Berestein G; Hung MC; Lupu R; Tari AM Oncogene 2000, 19, 6271. [DOI] [PubMed] [Google Scholar]
- 4.Caravatti G; Rahuel J; Gay B; Furet P Bioorg. Med. Chem. Lett 1999, 9, 1973. [DOI] [PubMed] [Google Scholar]
- 5.Long Y-Q; Yao Z-J; Voigt JH; Lung F-DT; Luo JH; Burke TR; King CR; Yang D; Roller PP Biochem. Biophys. Res. Commun 1999, 264, 902. [DOI] [PubMed] [Google Scholar]
- 6.Rahuel J; Gay B; Erdmann D; Strauss A; Garcia-Echeverria C; Furet P; Caravatti G; Fretz H; Schoepfer J; Grütter MJ Nat. Struct. Biol 1996, 3, 586. [DOI] [PubMed] [Google Scholar]
- 7.Ogura K; Tsuchiya S; Terasawa H; Yuzawa S; Hatanaka H; Mandiyan V; Schlessinger J; Inagaki FJ J. Biomol. NMR 1997, 10, 273. [DOI] [PubMed] [Google Scholar]
- 8.Ogura K; Tsuchiya S; Terasawa H; Yuzawa S; Hatanaka H; Mandiyan V; Schlessinger J; Inagaki FJ Mol. Biol 1999, 289, 439. [DOI] [PubMed] [Google Scholar]
- 9.Shakespeare WC Curr. Opin. Chem. Biol 2001, 5, 409. [DOI] [PubMed] [Google Scholar]
- 10.Machida K; Mayer BJ BBA-Proteins Proteomics 2005, 1747, 1. [DOI] [PubMed] [Google Scholar]
- 11.Muller G Top. Curr. Chem 2001, 211, 17. [Google Scholar]
- 12.Fretz H; Furet P; Garcia-Echeverria C; Schoepfer J; Rahuel J Curr. Pharm. Des 2000, 6, 1777. [DOI] [PubMed] [Google Scholar]
- 13.Oligino L; Lung F-DT; Sastry L; Bigelow J; Cao T; Curran M; Burke TR; Wang S-M; Krag D; Roller PP; King CR J. Biol. Chem 1997, 272, 29046. [DOI] [PubMed] [Google Scholar]
- 14.Li P; Peach ML; Zhang M; Liu H; Yang D; Nicklaus M; Roller PP Bioorg. Med. Chem. Lett 2003, 13, 895. [DOI] [PubMed] [Google Scholar]
- 15.Jiang S; Li P; Lai CC; Kelley JA; Roller PP J. Org. Chem 2006, 71, 7307. [DOI] [PubMed] [Google Scholar]
- 16.Jiang S; Li P; Peach ML; Bindu L; Worthy KW; Fisher RJ; Burker TR Jr.; Nicklaus M; Roller PP Biochem. Biophys. Res. Commun 2006, 349, 497. [DOI] [PubMed] [Google Scholar]
- 17.Song Y-L; Roller PP; Long Y-Q Bioorg. Med. Chem. Lett 2004, 14, 3205. [DOI] [PubMed] [Google Scholar]
- 18.Song Y-L; Peach ML; Roller PP; Qiu S; Wang S; Long Y-QJ Med. Chem 2006, 49, 1585. [DOI] [PubMed] [Google Scholar]
- 19.Li P; Zhang M; Peach ML; Zhang X; Liu H; Nicklaus M; Yang D; Roller PP Biochem. Biophys. Res. Commun 2003, 307, 1038. [DOI] [PubMed] [Google Scholar]
- 20.Lung F-DT; Long Y-Q; King CR; Varady J; Wu X-W; Wang S; Roller PP J. Peptide Res 2001, 57, 447. [DOI] [PubMed] [Google Scholar]
- 21.Lou Y-C; Lung F-DT; Pai M-T; Tzeng S-R; Wei S-Y; Roller PP; Cheng J-W Arch. Biochem. Biophys 1999, 372, 309. [DOI] [PubMed] [Google Scholar]
- 22.Mislow K; Green MM; Laur P; Melillo JT; Simmons T; Ternay AL Jr. J. Am. Chem. Soc 1965, 87, 1958. [Google Scholar]
- 23.Ottenheijm HCJ; Liskamp RMJ; Helquist P; Lauher JW; Shekhani MS J. Am. Chem. Soc 1981, 103, 1720. [Google Scholar]
- 24.Kubec R; Musah RA Phytochemistry 2001, 58, 981. [DOI] [PubMed] [Google Scholar]
- 25.Liskamp RMJ; Zeegers HJM; Ottenheijm HC J. J. Org. Chem 1981, 46, 5408. [Google Scholar]
- 26.Van den Broek LAGM; Breuer ML; Liskamp RMJ; Ottenheijm HC J. J. Org. Chem 1987, 52, 1511. [Google Scholar]
- 27.(a) Li P; Jiang S; Lee S-L; Lin CY; Johnson MD; Dickson RB; Michejda CJ; Roller PP J. Med. Chem 2007, 50, 5976; [DOI] [PubMed] [Google Scholar]; (b) Kamenecka TM; Danishefsky SJ Chem.-Eur. J 2001, 7, 41; [DOI] [PubMed] [Google Scholar]; (c) Kamenecka TM; Danishefsky SJ Angew. Chem., Int. Ed 1998, 37, 2993; [DOI] [PubMed] [Google Scholar]; (d) Kamenecka TM; Danishefsky SJ Angew. Chem., Int. Ed 1998, 37, 2995; [DOI] [PubMed] [Google Scholar]; (e) Jou G; González I; Albericio F; Lloyd-Williams P; Giralt EJ Org. Chem. 1997, 62, 354; [DOI] [PubMed] [Google Scholar]; (f) Jiang S; Li P; Lee S-L; Lin CY; Long Y-Q; Johnson MD; Dickson RB; Roller PP Org. Lett. 2007, 9, 9. [DOI] [PubMed] [Google Scholar]
- 28.GLIDE, Version 4.5; Schrodinger, LLC: New York, NY, 2007. [Google Scholar]
- 29.Friesner RA; Banks JL; Murphy RB; Halgren TA; Klicic JJ; Mainz DT; Repasky MP; Knoll EH; Shelley M; Perry JK; Shaw DE; Francis P; Shenkin PS J. Med. Chem 2004, 47, 1739. [DOI] [PubMed] [Google Scholar]
- 30.Halgren TA; Murphy RB; Friesner RA; Beard HS; Frye LL; Pollard WT; Banks JL J. Med. Chem 2004, 47, 1750. [DOI] [PubMed] [Google Scholar]
- 31.Friesner RA; Murphy RB; Repasky MP; Frye LL; Greenwood JR; Halgren TA; Sanschagrin PC; Mainz DT J. Med. Chem 2006, 49, 6177. [DOI] [PubMed] [Google Scholar]
- 32.MACROMODEL 9.5; Schrodinger, LLC: New York, NY, 2007. [Google Scholar]
- 33.Nioche P; Liu W-Q; Broutin I; Charbonnier F; Latreille M-T; Vidal M; Roques B; Garbay C; Ducruix AJ Mol. Biol 2002, 315, 1167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.