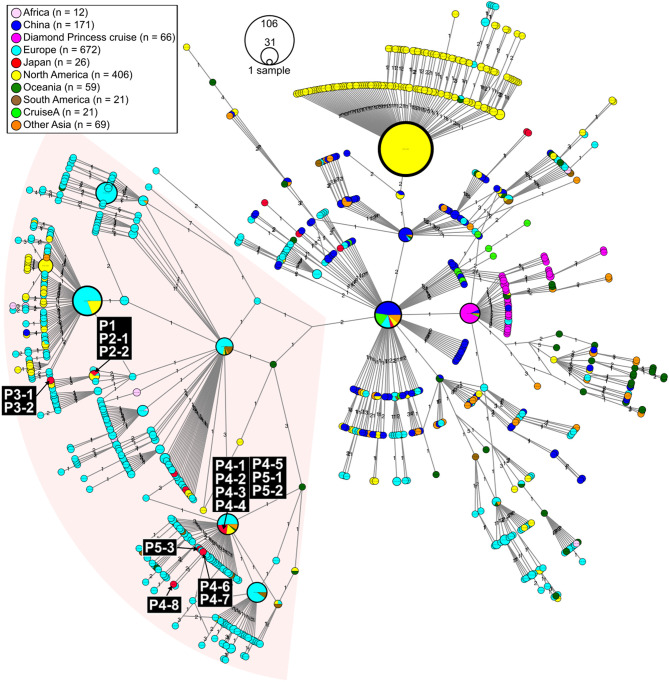
Figure 2.
Haplotype network using genome-wide single-nucleotide variations (HN-GSNVs) of SARS-CoV-2 isolates in the world. Whole-genome sequences of SARS-CoV-2 isolates from 10 travelers who returned from Egypt and possible patients linked to them (Figure 1) were compared with all GISAID-available SARS-CoV-2 genomes (n = 1,507, updated on March 30, 2020. See Table S1) by median-joining SNV network analysis. The numbers on the edges indicate differential SNVs between pair-wise nodes (isolates). SARS-CoV-2 disseminated from the end of December, 2019, from Wuhan City in China, one of the potential origins of Wuhan-Hu-1, isolated on December 26, 2020 (GenBank ID: MN908947). Wuhan-Hu-1 is plotted at the center of the haplotype network. Currently, at least three clades have disseminated globally in a region-specific manner.