Abstract
TP53 mutation in acute myeloid leukemia (AML) is associated with poor prognosis. Since no targeted therapy is available to restore p53 function, it is of great interest to test whether other pathways activated by TP53 mutations can be therapeutically targeted. Here we showed HIF-1α target genes are enriched in TP53-mutated vs TP53-wild type AML. To determine the role of this activation, we tested efficacy of HIF-1α inhibitor echinomycin in TP53-mutated AML samples in vitro and in vivo. Echinomycin was broadly effective against a panel of primary AML blast cells, with low nanomolar IC50s and, based on colony-forming unit assay, was 10-fold more effective in eliminating AML stem cells. Echinomycin selectively eliminated CD34+CD38- AML cells. To test the therapeutic efficacy of echinomycin, we established a xenograft model of TP53-mutated AML. Echinomycin was broadly effective against xenografts from multiple AML samples in vivo, and more effective than cytarabine + daunorubicin chemotherapy. Importantly, while cytarabine + daunorubicin enriched for AML stem cells, echinomycin nearly eliminated this population. Using TP53-mutated AML cell line THP1 and patient-derived AML cells, we tested a new echinomycin formulation with longer half-life and significantly improved therapeutic effect. Our data suggest a novel approach to treat AML with TP53 mutations.
Keywords: TP53, AML, HIF1α, echinomycin, liposome
Introduction:
Somatic TP53 mutations are frequently detected in a variety of cancers, with different frequencies dependent on the cancer type 1–3. TP53 mutations are found in acute myeloid leukemia (AML) patients with a frequency of over 10%, especially in cases with complex karyotypes 4–6, and are found at even higher frequencies in therapy-related AML (between 20–40%)7–9. Overall, TP53 mutations are associated with very poor prognosis, with poor responses to chemotherapy 5, 10, 11 and allogeneic stem cell transplantation 12. Response rates to hypomethylating agents are higher, but responses are not durable11, 13. Restoration of p53 function is a possible strategy to suppress cancer growth, but no targeted therapy is available clinically to restore p53 function. It is of great interest to test whether other pathways activated by TP53 mutations can be therapeutically targeted.
TP53 represses HIF-1-stimulated transcription, and loss of TP53 function in tumor cells has been shown to cause defective MDM2-mediated degradation of HIF-1α 14, 15. Accumulation of HIF-1α associated with loss of p53 function has been associated with an unfavorable prognosis and increased risk of patient mortality in some malignancies 16. We discovered that, even under normoxia, HIF1α signaling was selectively activated in the stem cells of murine T-ALL and human AML 17. Other studies have confirmed that the pathway is also critical for the maintenance of chronic myeloid leukemia stem cells and could be targeted to provide therapeutic effect 18, 19. Importantly, the HIF1α inhibitor echinomycin efficiently eradicated murine leukemia and was highly effective and selective in eliminating AML stem cells without adverse effect on normal hematopoietic stem cells 20, 21. Although TP53 mutations are observed in approximately 10% of AML samples and are associated with devastating prognosis 22, it has not been previously documented whether inhibition of HIF-1α accumulation in TP53-mutated AML suppresses leukemia cell progression. It is of great interest to test whether targeting the HIF-1α pathway can provide a therapeutically effective strategy for TP53-mutated AML.
Here, we found that HIF-1α target genes are enriched in TP53-mutated, vs TP53-wild type, AML and that echinomycin is broadly effective against multiple TP53-mutated AML samples in xenograft mouse models. We found that echinomycin is more effective than Daunorubicin+Cytarabine (DNR+Ara-C) therapy. Importantly, while DNR+Ara-C therapy enriched for AML stem cells, echinomycin largely eliminated this population both in vitro and in vivo. We developed a new echinomycin formulation with longer half-life and significantly improved therapeutic effect for TP53-mutated AML. Our data suggest a novel approach to treating AML with TP53 mutation.
Results
CD34+CD38- human primary TP53-mutated AML cells are more sensitive to echinomycin.
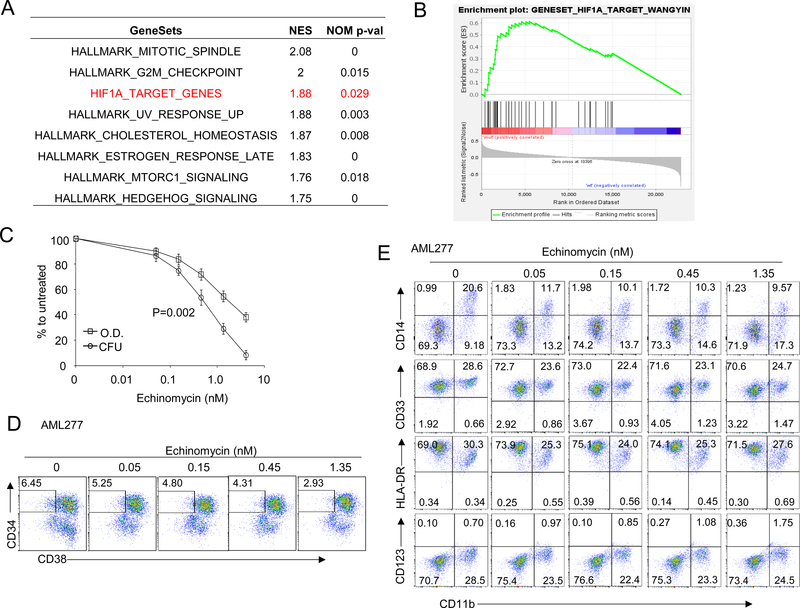
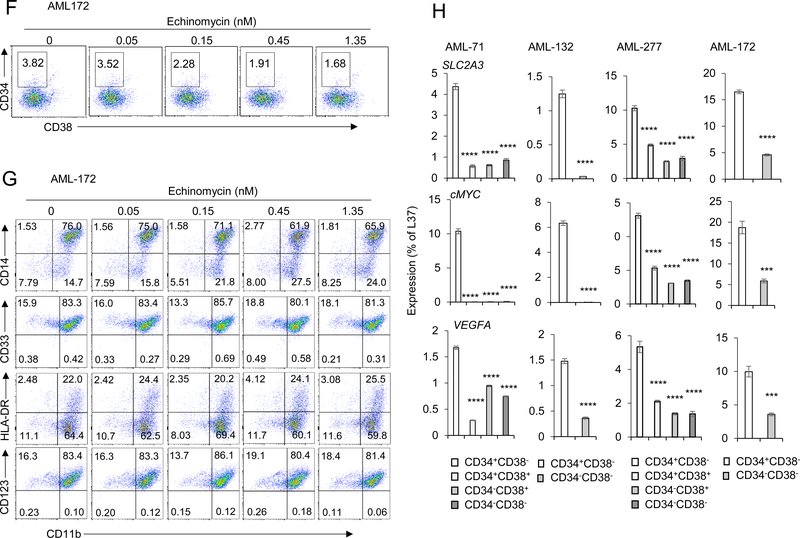
We analyzed a published dataset 23 by GSEA for gene sets that are significantly enriched in 32 AML samples harboring TP53 mutant compared to 419 patients TP53 wild type patients with RNA-seq data. Among 50 hallmark gene sets from Molecular Signatures Database (MSigDB), and using FDR<0.1 as the threshold, we identified 8 sets that were enriched in patients with TP53-mutated AML samples. Among them, HIF-1α target genes were most significantly enriched (Figure 1A, B). We also compared the expression of HIF-1α target genes in AML patients from The Cancer Genome Atlas (TCGA). Patients with TP53 mutation had enriched activity of HIF1A target genes compared to TP53 wild type patients, which was confirmed by the dramatic upregulation of eight HIF1A-target marker genes (Fig. S1). We tested the effect of echinomycin, an inhibitor of HIF-1α, on seven primary TP53-mutated AML samples by incubating the cells with 0.05 to 4.05 nM echinomycin and measuring cell viability and colony formation by MTT and colony-forming cell assays, respectively. Echinomycin had potent cytotoxic effects on TP53-mutated AML-277, with EC50 of 2.075 nM after only 24 hrs incubation time (Fig. 1C), and EC50 of about 0.5 nM if incubated for 48 hrs (Fig. S2). Compared to AML blasts, colony-forming unit (CFU) AML subsets were approximately 2.5-fold more sensitive to echinomycin, exhibiting EC50s of about 0.883 nM (Fig. 1C). We tested six additional TP53-mutated AML samples and found similar results in sensitivity to echinomycin in MTT and CFU, summarized in Table 1. The EC50s in the MTT assay are in the range of 0.656 nM to 3.021 nM, while EC50s in the CFU assay are in the range of 0.113 to 0.833 nM (Table 1). These data demonstrated a critical role for HIF-1α in CFU activity in seven primary TP53-mutated AML samples. To test whether echinomycin selectively targeted subsets of the TP53-mutated AML cells in the CFU assay, we analyzed the percentage of CD34+CD38- cells from the colonies on day 10 after treatment with echinomycin. CD34+CD38- AML subsets, which are leukemia stem cells, were decreased in a concentration-dependent manner in primary TP53-mutated AML, and were decreased more than two-fold in the colonies treated with 1.35 nM of Echinomycin compared to vehicle-treated cells (Fig. 1D, F). However, there was no significant difference in the frequency of subsets with other leukemia markers (Fig. 1E, G). These data suggested that HIF-1α activity is required more for maintenance of the CD34+CD38- AML subset compared to bulk leukemia cells. We previously reported that enrichment of HIF-1α in TP53 wild type CD34+CD38- AML stem cells formed the basis for their selective elimination by echinomycin 20. To test if this was also the case for TP53-mutated AML, we compared mRNA levels of HIF-1α target genes among sorted CD34+CD38-, CD34-CD38-, CD34-CD38+, or CD34+CD38+ subsets for four clinical AML samples: AML 277 and AML 172 (TP53-mutated) and AML 71 and AML 132 (TP53-wild type), and the primer sequences are listed in Supplemental Table S1. Consistent with the bioinformatics data presented in Fig. 1A & B, our quantitative-PCR (qPCR) analysis revealed that levels of HIF-1α targets GLUT, cMYC and VEGF were altogether higher in TP53-mutated samples vs –wild type samples, regardless of the population analyzed. Notably, however, the expression of these targets was significantly higher in the CD34+CD38- stem cell subsets compared to the remaining fractions, regardless of TP53 mutational status (Fig. 1H). Taken together, the data indicated that HIF-1α plays a critical role in the CD34+CD38- stem cell subset of TP53-mutated AML, and that the elevated HIF-1α activity compared to other subsets underlies the sensitivity of these cells to echinomycin. Importantly, the data suggested that targeting HIF-1α by echinomycin might offer a therapeutic advantage in TP53-mutated AML in vivo, which is generally refractory to standard chemotherapies.
Figure 1. Echinomycin significantly inhibits expansion of human TP53-mutant AML cells in vitro.
(A&B) HIF-1α target genes are highly expressed in TP53-mutated AML from patients. Gene Set Enrichment Analysis (GSEA) was performed to examine the expression differences of HIF-1α targets between patients with TP53-mutated AML and TP53-wild type AML from the public database. GSEA result showing significant enrichment (FDR<0.1) of the curated HIF-1α target genes and hallmark gene sets from MSigDB in TP53-mutated AML patients (32 cases) compared with patients with TP53-wild type AML (419 cases). In silico analysis of data reported by Tyner et al 23. (C) Sensitivity to echinomycin of TP53-mutated AML cells from patients. AML cells were incubated for 24 hours in culture medium with echinomycin at increasing concentrations, ranging from 0.05 to 4.05 nM. After the incubation period, MTT metabolization of treated cells was measured by absorbance and is expressed as the mean ± SD (n = 3) of the percentage of the value of untreated cells. At the same time, 2X104 treated cells were seeded in methylcellulose medium for colony-forming assay. Colonies were counted on day 7 to 10 in different AML cells. CFU is expressed as the mean ± SD (n = 3) of the percentage of the value of untreated cells. Data are shown for one experiment and are representative of two independent experiments. (D-G) Selectivity of echinomycin for CD34+ CD38- subset of TP53-mutated AML cells. Cells from colonies as described in Fig.1B were resuspended and stained with anti-human CD45, CD34, CD38, CD14, CD33, HLA-DR, CD11b and CD123, and analyzed by FACS. Representative FACS plots show the percentage of human CD34+ CD38- cells (D, F) or cells with leukemia markers (E, G) in colonies treated with echinomycin. (H) CD34+CD38- AML subsets have higher HIF1α activity compared to bulk tumor cells. Two TP53-mutated (AML 277, AML 172) and two TP53-wild type (AML 71, AML 132) clinical AML samples were sorted into CD34+CD38-, CD34-CD38-, CD34-CD38+, or CD34+CD38+ fractions by FACS and qPCR was subsequently performed to analyze mRNA levels of HIF-1α target genes among each subset. The data are summarized and statistical significance was determined by unpaired t test of the CD34+CD38- cells vs the other subsets (* p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001; ns, not significant). All data are representative of two independent experiments.
Table 1.
IC50 of MTT and CFU (Mean ± SD) of TP53-mutated and -wild type AML cells treated with Echinomycin
| TP53-mutated AML | TP53-wild type AML | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| AML-12 | AML-83 | AML-135 | AML-147 | AML-172 | AML-253 | AML-277 | AML-71 | AML-132 | |
| IC50 (MTT, nM) | 3.021 ± 1.145 | 0.656 ± 0.061 | 2.017 ± 0.205 | 1.252 ± 0.325 | 2.405 ± 0.344 | 1.274 ± 0.148 | 2.075 ± 0.213 | 3.438 ± 1.295 | 3.276 ± 0.979 |
| IC50 (CFU, nM) | 0.152 ± 0.021 | 0.113 ± 0.067 | 0.232 ± 0.061 | 0.237 ± 0.014 | 0.376 ± 0.121 | 0.326 ± 0.078 | 0.833 ± 0.039 | 0.184 ± 0.179 | 0.175 ± 0.098 |
Echinomycin impairs leukemia progression in mice xenografted with primary TP53-mutated AML
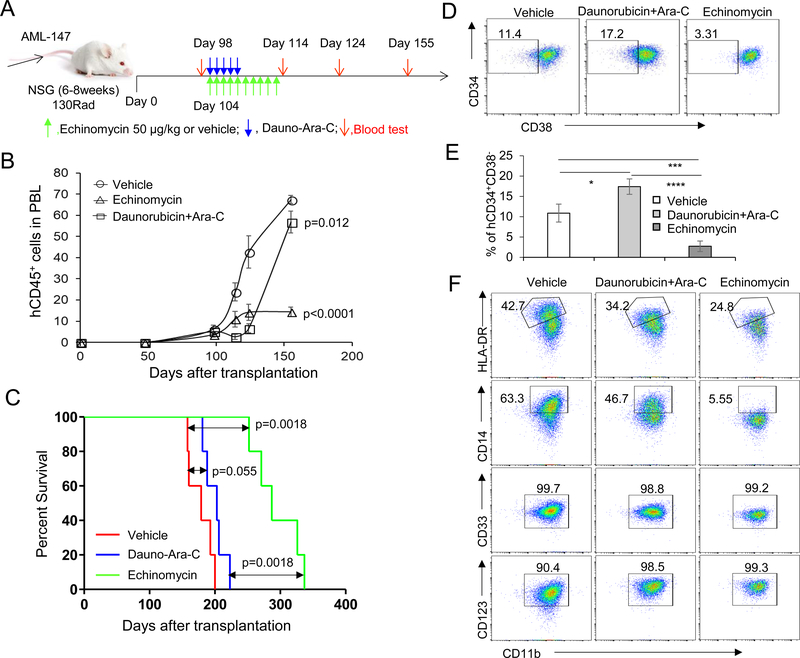
To explore the therapeutic potential of echinomycin in vivo, mice were grafted with primary human TP53-mutated AML cells (AML-147) and treated with echinomycin or conventional chemotherapy consisting of DNR+Ara-C, according to the dosing schedule in Fig. 2A. Following i.v. transplantation of 1X106 TP53 mutant AML-147 cells into 1.3 Gy-irradiated NOD/SCID IL2rgnull (NSG) recipients, we monitored the reconstitution and progression of the cells by FACS analysis of human CD45+ cells in the peripheral blood. By day 98 after transplantation, between 2 to 10% of the peripheral blood cells were human CD45+, and we initiated treatment with Echinomycin or DNR+Ara-C on day 104. Upon completion of the treatment cycle, FACS analysis of the peripheral blood indicated that the growth of TP53-mutated AML cells was significantly inhibited in recipients either treatment. However, the leukemia cells relapsed in DNR+Ara-C-treated mice after cessation of treatment due to dose-limiting toxicity. In contrast, echinomycin-treated mice exhibited a prolonged period of growth inhibition, with peripheral blood AML blasts remaining below 20% when vehicle-treated mice had blast levels exceeding 70% and started dying (Fig. 2B). Similarly, mice treated with either drug regimen experienced significantly prolonged survival times vs vehicle-treated mice, but survival was more prolonged for echinomycin-treated mice (Fig. 2C). These data demonstrated that HIFs may serve as an effective therapeutic target for TP53-mutated AML.
Figure 2. Echinomycin significantly suppresses expansion of human TP53-mutated AML cells in a mouse xenograft model.
(A) Experimental design for the AML-147 xenograft model. NSG mice were irradiated with 1.3 Gy and transplanted with 1X106 human TP53-mutated AML-147 cells by i.v. injection (day 0). Peripheral blood tests were performed before drug treatment on day 98, and after treatment on days 114, 124, and 155, indicated by red arrows. Drugs were administered by i.v. injection beginning on day 104. Individual doses are indicated by green arrows for echinomycin (50 μg/kg) or blue arrows for conventional therapy. The regimen for conventional therapy consisted of cytarabine (5 mg/kg) + daunorubicin (0.5mg/kg) for 3 continuous days, then daunorubicin (0.5 mg/kg) for 2 continuous days. (B) Human CD45+ cells in blood of AML-147 recipients described in (A) were detected by FACS analysis on day 98 after transplantation. Frequencies of human CD45+ in blood of recipients before treatment (Day 98) and after echinomycin treatment (Day 114, 124 and 155) are summarized. Statistical analysis was performed by one-way ANOVA with Tukey’s multiple comparisons test, p values are shown for either drug treatment group vs vehicle control. (C) Kaplan–Meier survival curves of AML-147 recipients in (A) treated with echinomycin, DNR+Ara-C, or vehicle. Data are representative of two independent experiments. (D-F) CD38-CD34+ subset from TP53-mutated AML cells are more sensitive to Echinomycin in vivo. Single cell suspensions isolated from spleens of mice described in (A) were stained with anti-human CD45, CD34, CD38, CD14, CD33, HLA-DR, CD11b and CD123, and analyzed by FACS. Representative FACS plots showing the percentage of human CD38-CD34+ cells in splenocytes from mice treated with Echinomycin, DNR+Ara-C, and vehicle are shown in (D) and summarized in (E), and representative FACS plots are shown for various leukemia marker expression patterns among the groups in (F). N=3 mice per group for vehicle and DNR+Ara-C groups, n=5 for echinomycin group, and statistical significance was determined by unpaired ttests between groups (* p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001; ns, not significant). All data are representative of two independent experiments.
Notably, while the frequencies of CD34+CD38- AML subsets decreased in echinomycin-treated mice in comparison to that in the vehicle-treated mice, they were significantly increased in DNR+Ara-C-treated mice (Fig. 2D&E). We also examined the frequencies of other AML cell subsets but found no significant difference between the three groups, except for a significant reduction in HLA-DRhiCD11b+ cells in echinomycin-treated mice (Fig. 2F). Collectively, these data suggest that CD34+CD38- AML stem cell subsets are especially sensitive to echinomycin. In contrast, DNR+Ara-C combination presumably targeted differentiated AML blasts and enriched for CD34+CD38- AML subsets, resulting in recurrence after cessation of DNR+Ara-C treatment.
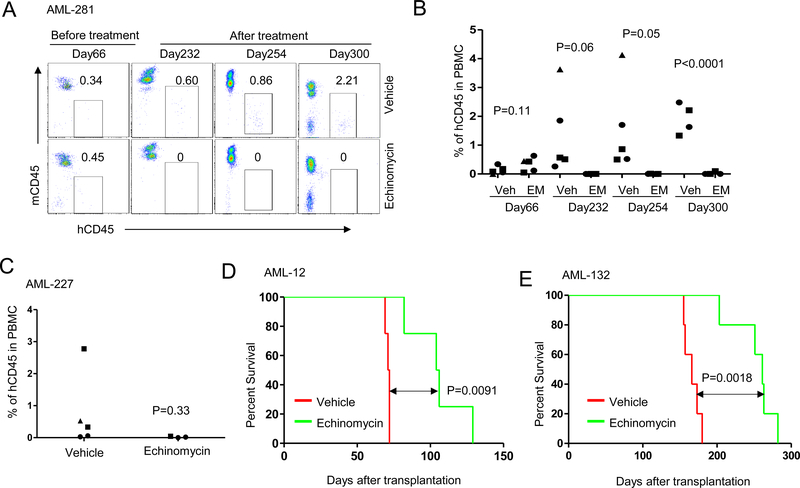
To further investigate the therapeutic effects of echinomycin in the context of TP53 mutations, we performed similar analyses in four additional xenograft models, including three primary TP53-mutated samples (AML-281, AML-227, AML-012) and one primary TP53-wild type sample (AML-132). As shown in Figures 3A&B, by day 66 following transplantation of AML-281 cells, approximately 0.5% human CD45+ cells could be detected on average in the peripheral blood of the mice. Starting on day 67, we treated the AML-281 recipients with echinomycin 10μg/kg in a regimen consisting of three QDx5 cycles, each separated by two days of rest. FACS analysis of the peripheral blood cells showed that while the human CD45+ cells increased in vehicle-treated mice over the course of the study, they were undetectable in 100% of echinomycin-treated mice by day 232, and remained undetectable in nearly all of the echinomycin-treated mice until the end of the observation period on day 300 (Fig 3A&B). Using the same treatment regimen, we performed this experiment on mice xenografted with TP53-mutated AML-227 cells and found that human CD45+ cells were similarly reduced in echinomycin-treated mice compared to the vehicle group on day 300 (Fig. 3C). Due to the slow growing nature of AML-281 and AML-227 primary cells in the NSG recipients, mortality was not a feasible endpoint for the studies involving these two samples. In contrast, transplantation of primary TP53-mutated AML-012 cells resulted in mortality in 100% of NSG recipients by day 75 after transplantation in the absence of therapeutic intervention; accordingly, echinomycin extended survival time in AML-012 recipients by more than 130 days (Fig. 3D). As our initial findings presented in Fig. 1 revealed elevated HIF-1α activity in TP53-mutated AML, we sought to determine if differences in TP53 genotype might correspond to an overall increase in sensitivity to echinomycin. As summarized in Table 1, in vitro treatment of AML-71 and AML-132 with echinomycin revealed modestly increased IC50s for these cells when compared with multiple TP53 mutated samples, whereas the CFU indicated no difference in sensitivity to echinomycin. To test the impact of TP53 mutations on therapeutic effect of echinomycin in the xenograft models, we transplanted NSG mice with TP53-wild type AML-132 cells and observed the effects of echinomycin treatment on survival time for comparison with the TP53-mutated models. Although these cells exhibited lower levels of HIF-1α activation compared to the TP53-mutated samples, Echinomycin also significantly prolonged the survival time of the AML-132 recipients (Fig 3E). On the basis of these findings, it is evident that TP53-mutated AML clearly exhibits higher overall HIF-1α activity compared to cases of TP53-wild type AML, although the increased activity does not necessarily equate to an obvious increase in therapeutic response to echinomycin. Rather, the results suggest that a basal requirement for HIF-1α pathway activation in the CD34+CD38- stem cell subset, which we uncovered previously in cases of TP53-wild type AML, appears to be indispensable also in cases with TP53 mutation. Nevertheless, the finding that TP53 mutations did not abrogate therapeutic response to Echinomycin is very significant as they have rendered leukemia more resistant to conventional therapy.
Figure 3. Echinomycin has therapeutic efficacy in most human TP53-mutated AMLs.
(A) NSG mice irradiated with 1.3 Gy were given i.v. injection of 1X106 human primary TP53-mutated AML cells (AML-281). Human CD45+ cells in blood of recipients were detected by FACS analysis on day 66 after transplantation. Starting on day 67, mice received 10 μg/kg of echinomycin or vehicle by i.p. injections on a schedule consisting of 3 QDx5 cycles, each separated by 2 days rest. The FACS file showed the percentage of human CD45+ in blood of recipients before treatment (Day 66) and after echinomycin treatment (Day 232, 254 and 300) in two representative mice (vehicle and echinomycin treatment). (B) Summary of percentages of human CD45+ in blood of all recipients treated with vehicle or echinomycin. Data are representative of two independent experiments. (C) NSG mice were irradiated with 1.3 Gy and given 1X106 human primary TP53-mutated AML cells (AML-227) by i.v. injection and were treated with echinomycin according to the schedule in A. Summary of percentages of human CD45+ in blood of all recipients treated with vehicle or echinomycin on day 300 is shown. Data are representative of two independent experiments. (D) NSG mice were irradiated with 1.3 Gy and given i.v. injection of 1X106 human TP53-mutated AML cells (AML-012). Twenty days later, mice received 50 μg/kg of echinomycin or vehicle by i.p. injection on a schedule consisting of Q2Dx10. Kaplan–Meier survival curves of NSG recipients treated with echinomycin and vehicle are shown. Data are representative of two independent experiments. (E) NSG mice were irradiated with 1.3 Gy and given i.v. injection of 2X106 human TP53-wild type AML cells (AML-132). Ninety-three days later, the mice were treated with echinomycin (50 μg/kg) or vehicle, Q2Dx10, by i.p. (n=5/group). Kaplan–Meier survival curves of NSG recipients treated with echinomycin and vehicle are shown. Data are representative of two independent experiments.
A liposomal formulation of echinomycin significantly prolonged the survival of TP53-mutated AML xenografts
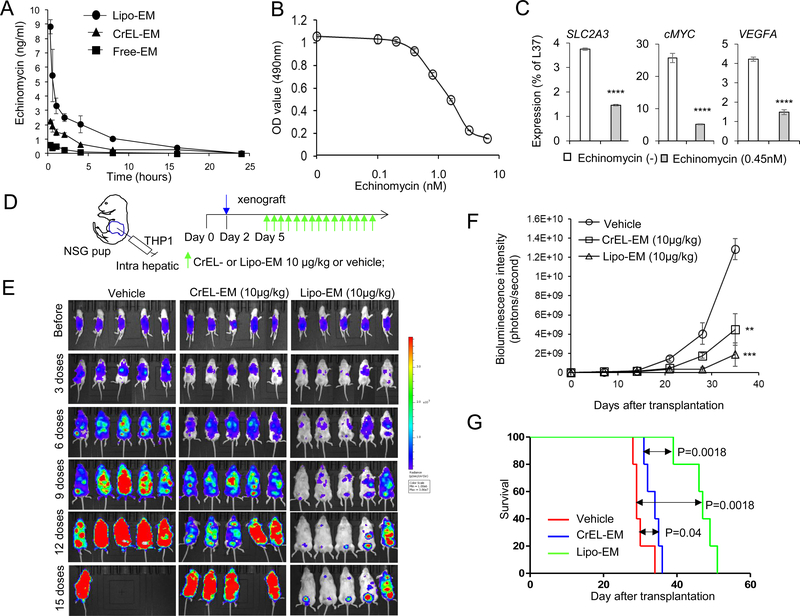
Pharmacokinetics (PK) of echinomycin in vivo have not been studied due to the lack of a sensitive analytical method for assaying echinomycin concentrations in biological matrices. To overcome this challenge, we developed and validated a sensitive LC-MS/MS method for in vivo detection of echinomycin, which we used to study the PK and tissue distribution of echinomycin in mice. As shown in Fig. S3, the new method provided a lower limit of quantification (LLOQ) for echinomycin in plasma and tissue samples of 50 pg/mL (pg/g). We studied the plasma PK of 3 echinomycin formulations following a single i.v. injection at a dose of 100 ug/kg. As shown in Fig. 4A, free echinomycin, consisting of DMSO/saline (1:9, v/v), reached plasma concentrations of 0.61, 0.51, 0.11 and 0.063 ng/ml at 0.25, 1, 4 and 8 hrs following administration, respectively. At the same respective time points, plasma echinomycin concentrations reached 2.3, 1.5, 0.64 and 0.28 ng/ml when formulated in Cremophor EL/ethanol/saline (1:1:18, v/v) (CrEL-EM), and 8.9, 3.0, 2.1 and 1.0 ng/ml when formulated in liposomes consisting of HSPC:Cholesterol:DSPE-mPEG2000 (57:38:5, mol:mol) at a 3% drug/lipid molar ratio. The results demonstrated that formulating echinomycin in PEGylated liposomes significantly prolonged the circulation time in the bloodstream compared to the alternative formulations. Table 2 shows the mean values for the pharmacokinetic parameters in plasma after single doses of the three formulations of echinomycin administered to mice. Echinomycin exposure for the liposomal formulation was much higher than for free drug and CrEL-EM. The estimated AUClast of the liposomal formulation was 13.6-and 3.19-fold that of free drug and CrEL-EM, respectively. For echinomycin Cmax in mouse plasma, the liposomal formulation was 13.2 and 3.90 times higher than for free drug and CrEL-EM. In addition, liposomal formulation and CrEL-EM showed a similar elimination half-life (T1/2) which was around twice as long as for free drug. Taken together, the liposomal formulation showed the best pharmacokinetic behavior in mice among these three formulations.
Figure 4. Liposomal echinomycin suppressed the growth of human TP53 null THP1 cells and patient-derived xenograft TP53-mutated AML 12 cells in xenograft mouse model.
(A) Pharmacokinetic study of echinomycin in plasma of mice. NSG mice were given a single i.v. injection of control vehicle or echinomycin at 100 ug/kg in one of three different formulations (Free-EM: echinomycin dissolved in DMSO then dispersed in PBS; CrEL-EM, echinomycin dispersed in cremophor; Lipo-EM, liposomal echinomycin). Blood samples were collected at 0.25, 0.5, 1, 2, 4, 8, 16 and 24h after dosing. The concentration of echinomycin in plasma was analyzed by LC-MS/MS method. (B) Echinomycin inhibits the growth of TP53-null THP1 cells. THP1 cells (1X105) were seeded in RPMI 1640 culture medium in a 24-well plate and cultured for 24 hours. The cells were treated with echinomycin at different concentrations dissolved in DMSO (free echinomycin) for 48 hrs. MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] (10μl of 5mg/ml) was added to each well containing THP1 cells. After 2 to 4 hours in culture, cells were centrifuged and the formazan crystals were resuspended in 150 μl DMSO and optical density was read at 490 nm. The values for the measured wells after background subtraction are summarized. Data shown are means and S.D. of triplicate wells and are representative of three independent experiments. (C) Quantitative-PCR analysis of mRNA isolated THP1 cells cultured with vehicle or echinomycin (0.45 nM) for 24 hrs. Statistics are by paired student’s t test for wells analyzed in triplicate per each group (* p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001; ns, not significant). Data are representative of two independent experiments. (D) Dosing regimen of echinomycin treatment for mice transplanted with luciferase-transduced THP1 cells. Day 0 indicates the date of birth and 1X106 of THP1 cells are transplanted into pups via intrahepatic injection on day 2 (blue arrow). The baseline pre-treatment bioluminescence is determined by imaging the mice on day 5. Mice were treated with 10 μg/kg CrEL-EM or liposomal echinomycin, or vehicle, according to a Q2Dx15 schedule starting on day 3 after tumor transplantation. (E) Therapeutic effect of echinomycin. Serial imaging was performed for echinomycin-or vehicle-treated NSG recipients of THP1 as described in (D). Imaging is shown for each group on day 5, corresponding to the pre-treatment values (before), and on days 3, 6, 9, 12 and 15 after doses. Data are representative of 3 independent experiments. (F) Quantification of bioluminescence intensity of mice depicted in (E). Bioluminescence intensity (photons/second) was measured and plotted before and after treatment and is shown as means ± SEM (n=7 per group). Statistics are by t test (* p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001; ns, not significant). (G) Kaplan-Meier survival curves are shown for the mice as described in (D). Liposomal echinomycin had significantly prolonged survival compared with vehicle treatment. Data are representative of two independent experiments.
Table 2.
PK parameters (Mean ± SD) of Echinomycin in mouse plasma after administration of 0.1 mg of three formulation of Echinomycin
| T1/2 (hr) | Cmax (ng/mL) | AUClast (hr*ng/mL) | |
|---|---|---|---|
| Free-EM | 2.46 ± 0.29 | 0.67 ± 0.24 | 1.78 ± 0.55 |
| CrEL-EM | 4.68 ± 0.65 (P=0.0057)a) | 2.27 ± 0.11 (P=0.00047) | 7.58 ± 0.45 (P=0.00015) |
| Lipo-EM | 4.28 ± 0.61 (P=0.0095) | 8.85 ± 0.88 (P=0.0001) | 24.17 ± 2.58 (P=0.00012) |
P value to Free-EM.
Echinomycin inhibits expansion of human TP53-null THP1 cells in xenografted mouse model.
THP1 is an acute monocytic leukemia cell line that has a homozygous 26-base deletion starting at codon 174 of the TP53 coding sequence24. We first investigated the therapeutic effect of echinomycin on TP53-null THP1 cells in vitro. THP1 cells were incubated for 48 hrs with 0.1–6.4 nM echinomycin and cell viability was determined by MTT assay (Fig. 4B). The THP1 cells exhibited a concentration-dependent decrease in cell viability in response to echinomycin treatment, with an EC50 of 1.25 nM (Fig. 4B) as well as suppression of HIF-1α target genes (Fig. 4C).
To evaluate the therapeutic effect of liposomal echinomycin against TP53-mutated AML cells, we transplanted THP1 into newborn NSG pups via intrahepatic injection (Fig. 4D). Mice were treated with 10 μg/kg of echinomycin formulated in either CrEL or liposomes on a QDx15 schedule beginning on day 3 after THP1 cell transplantation (Fig. 4D). Mice were imaged on day 3 after THP1 cell transplantation (before) and throughout the treatment cycle. In vehicle-treated mice, the average bioluminescence signal intensity increased over the course of three weeks after transplantation, indicating progressive leukemia growth, and all vehicle-treated mice died from leukemia within 35 days (Fig. 4E to G). In contrast, mice treated with echinomycin displayed reduced leukemia growth, indicating a therapeutic effect of echinomycin. CrEL-EM modestly inhibited bioluminescence signal intensity and provided a marginal, albeit significant, improvement in survival time; on the other hand, the bioluminescence signal barely increased during the same time period in mice treated with liposomal echinomycin, and these mice survived for more than 50 days (Fig. 4E to G). The survival of recipients of liposomal echinomycin was significantly longer vs mice that received vehicle or CrEL-EM (Fig. 4G). These results indicated a robust therapeutic effect of liposomal echinomycin against THP1 leukemia cells in vivo and demonstrate a significant anti-leukemia effect of liposomal echinomycin in TP53-null leukemia cells.
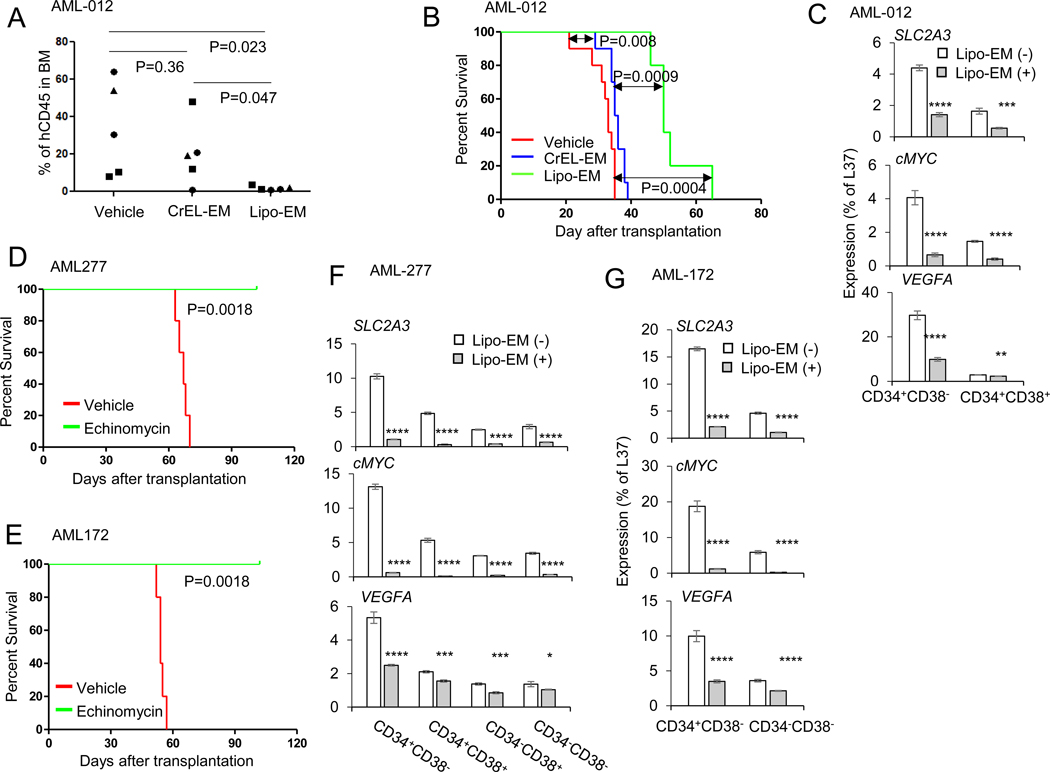
Liposomal echinomycin suppressed the growth of human patient-derived xenograft TP53-mutated AML cells in xenograft mouse model.
To further examine the therapeutic effect of liposomal echinomycin using patient-derived AML cells, we transplanted NSG mice with AML-012 and treated the mice with either CrEL or liposomal echinomycin, Q3Dx5 starting day 10 after transplantation to observe the effects of treatment using different formulations. Unlike other AML cells we used, AML-012 could not be detected in peripheral blood throughout the study period. Therefore, to observe the effects of the echinomycin treatment on the AML-012 blast growth at a fixed time point, we sacrificed 5 mice from each group on day 30 and analyzed human blasts in the BM. Liposomal echinomycin, but not CrEL-echinomycin, suppressed AML-012 blasts in the BM in this xenograft model (Fig. 5A). The survival of remaining mice in each group was significantly longer vs mice that received vehicle or CrEL-EM (Fig. 5B). To observe the pharmacodynamic activity of liposomal echinomycin on the HIF-1α pathway among different AML subsets, we examined levels of HIF-1α target genes in the sorted human CD34+CD38- or CD34+CD38+ AML-012 blasts isolated from BM of xenograft recipients of either vehicle or liposomal echinomycin. As expected, the levels of HIF-1α target genes were significantly reduced in the AML-012 blasts, regardless of the cell subset analyzed, although the target gene suppression was generally more profound among the CD34+CD38- cells (Fig. 5C). We performed similar experiments in additional TP53-mutated AML primary cells. As shown in Fig. 5D and E, liposomal echinomycin treatment was effective in extending the survival time of mice xenografted with AML-277 or AML-172 primary cells, respectively. Accordingly, HIF-1α target genes were also reduced in the sorted AML blasts isolated from the BM of liposomal echinomycin-vs vehicle treated mice in both of these models, and the suppression was similarly most profound in the CD34+CD38- subsets (Fig. 5F, G). Altogether, these results further indicated a robust therapeutic effect of liposomal echinomycin against patient-derived leukemia cells in vivo and demonstrate a significant anti-leukemia effect of liposomal echinomycin in TP53-mutated human leukemia cells.
Figure 5. Liposomal Echinomycin suppressed the growth of human patient-derived xenograft TP53-mutated AML cells in xenograft mouse model.
(A) Liposomal Echinomycin, but not CrEL-EM, suppressed BM AML-012 blasts in the xenograft mouse model. NSG mice were transplanted with TP53-mutated AML-012 (twice passaged in NSG mice) via i.v. and treated with vehicle, CrEL-EM (0.1 mg/kg) or Lipo-EM (0.35 mg/kg) via i.v., once every three days for a total of 5 doses, beginning on day 10 after transplantation. The shorter survival times for all mice is due to the increased aggressiveness of the cells after two passages in the NSG. Summary of hCD45+ blasts in BM shown. Statistics calculated by unpaired t test. (B) NSG mice were transplanted with TP53-mutated AML-012 and treated with Vehicle, CrEL-EM (0.1 mg/kg) or Lipo-EM (0.35 mg/kg) as in (B) and Kaplan–Meier survival curves of the mice are shown. (C) NSG mice were transplanted with TP53-mutated AML-012 and treated with Vehicle or Lipo-EM (0.35 mg/kg) once every three days for 3 doses, then sacrificed for qPCR analysis of mRNA isolated from the sorted BM cells pooled from n=3 mice per group. Statistics are by t test (* p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001; ns, not significant). Data are representative of two independent experiments. (D-E) NSG mice were transplanted with TP53-mutated AML-277 or TP53-mutated AML-172 and treated with Vehicle or Lipo-EM (0.1 mg/kg) once every other day for 10 doses. Kaplan–Meier survival curves of the mice xenografted with AML-277 (D) or AML-172 (E) are shown. (F-G) NSG mice were transplanted with TP53-mutated AML-277 or TP53-mutated AML-172 and treated with Vehicle or Lipo-EM (0.1 mg/kg) once every other day for 3 doses. Then, the mice were euthanized and qPCR of HIF1α targets was performed on the mRNA isolated from the sorted BM cells pooled from n=3 mice per group for either AML-277 (F) or AML-172 (G). Statistics are by t test (* p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001; ns, not significant). Data are representative of two independent experiments.
Discussion
TP53 mutations have emerged as one of the most common driver mutations in human cancer 3, 25 and thus should, in theory, be one of the most attractive targets for cancer therapy 26. However, due to the broad scope of inactivation mutations, restoration of p53 function has not been successful in the clinic. As an alternative approach, it is of great interest to identify druggable pathways that are selectively activated after TP53 mutation. Here we showed, by in silico analysis of an RNAseq database, that HIF-1α pathway is activated in TP53-mutated AML, as its targets are coordinately increased. More importantly, we have shown that echinomycin, which inhibits HIF-1a activity by binding to the promoter region of its target genes 27, not only kills TP53-mutated AML blasts, but is even more active against the AML stem cell population. This conclusion is based on both in vitro and in vivo models using multiple established AML cell lines and primary AML samples and by comparing with conventional chemotherapy treatment in a xenograft model established from a primary AML sample. These data provide compelling evidence that echinomycin, arguably the most potent HIF-1a inhibitor, with picomolar IC50 in vitro 17, may offer a new approach for unmet medical needs of patients with TP53-mutated leukemia.
Echinomycin is a member of the quinoxaline family originally isolated from Streptomyces echinatus in 1957 28. Echinomycin was never tested in human hematological malignancies until we identified its function in the treatment of human AML, targeting cancer stem cells 17. Although echinomycin was used in several phase II trials at a dose of 1200 μg/m2 in humans, no pharmacokinetic (PK) data emerged since no method was available to measure drug concentration. A critical barrier to drug development was the lack of a sensitive method to measure trace amounts of echinomycin in blood and in tissues for pharmacokinetic studies for dose optimization. Here, we developed a sensitive and specific LC-MS/MS assay to measure echinomycin in plasma that is capable of detecting 0.025 ng/ml in mouse plasma or tissue extracts. This should greatly improve future efforts to develop what is likely the most effective inhibitor of the HIF pathway for cancer therapy.
Long-circulation nanoparticles have significant clinical impact since they offer sustained systemic delivery and increased drug residence time in the bloodstream 29. Polyethylene glycol (PEG)-modified liposomes have been successfully used to prolong the circulation time of therapeutic agents in the bloodstream compared to free drug or non-PEGylated liposome formulations 30, 31–33. In addition, the use of saturated phospholipids along with high cholesterol content is known to improve the stability of liposomal drug formulations and prolong circulation time. We used one of the most successful PEGylated liposomal formulations for increasing drug circulation time, consisting of HSPC:cholesterol:DSPE-mPEG2000 (57:38:5, mol:mol), to increase the circulation time of echinomycin in the bloodstream, with more than 7-fold increase in total exposure within 24 hours. Importantly, our preliminary analysis of safety in mice showed that liposomal echinomycin used herein has significantly less toxicity despite increased exposure (data not shown). We believe that these data suggest that by releasing echinomycin over an extended period, liposomes reduced exposure of normal tissues to toxicity associated with high concentrations of echinomycin within very short period after dosing. This will not only increase drug availability for treatment of hematological malignancies, but also increase safety. By establishing the therapeutic effect of echinomycin in a clinically viable formulation, our data reported herein support further clinical development of echinomycin for treatment of the AML subtype with poorest prognosis.
Experimental Procedures:
Mice
All procedures involving experimental animals were approved by the Institutional Animal Care and Use Committee of the University of Maryland and the Children’s National Medical Center, where this work was performed. Male and female Nod.Scid.Il2rg0 (NSG) mice aged 6–8 weeks were purchased from the Jackson Laboratory.
AML Samples
Samples from AML patients diagnosed at the University of Michigan Comprehensive Cancer Center and the University of Maryland School of Medicine were used in this study. The studies were approved by the University of Michigan and University of Maryland School of Medicine Institutional Review Boards (IRBs). Written informed consent was obtained from all patients prior to sample procurement. We used World Health Organization (WHO) AML diagnostic criteria (>20% myeloblasts in the bone marrow or peripheral blood) and determined WHO subclassification through review of data from the time of diagnosis. The clinical characteristics of patients with AML are listed in Supplemental Table 2. THP1 cells were purchased from ATCC and tested negative for mycoplasma contamination.
Cell viability and methylcellulose colony formation assay
For the cell viability assay, AML cells (1×105/well) were seeded in a 24-well plate and cultured for 24 hours prior to treatment with echinomycin at different concentrations, and then incubated for 24 or 48 additional hours. Ten μl of 10 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma) in phosphate-buffered saline was added to each well. Plates were returned to the cell culture incubator for 2 to 4 hours. The resulting MTT formazan crystals were pelleted and dissolved by adding 150 μl DMSO. Absorbance was measured at a wavelength of 490 nm. The methylcellulose colony formation assay was performed according to the manufacturer’s recommendations (STEMCELL Technologies Inc., Vancouver, BC, Canada). Briefly, after 24 or 48 hour incubation with echinomycin, the aforementioned treated AML cells were washed with culture medium and seeded at 2×104/well in a 24-well plate. On day 7 to 10 after AML cells were seeded, colonies were counted and analyzed by FACS. Experiments were performed in triplicate.
Flow Cytometry
Blood was collected by submandibular bleeding at different times before and after AML transplantation. Fluorochrome-labeled antibodies were directly added into whole blood. After 30 min of staining, the stained samples were treated with FACS Lysing Solution to lyse the red blood cells and then washed to remove excess antibodies and debris. Spleens were gently ground with frosted objective slides and the cells were passed through a nylon cell strainer, washed three times with RPMI 1640, labeled with antibodies and analyzed for the presence of different human cell populations. Antibodies used were FITC-conjugated mouse anti-human CD123, PE-Cy7 conjugated mouse anti-human HLA-DR, PerCP-conjugated mouse anti–human CD33, BV421-conjugated mouse anti-human CD34 and anti-human CD11b, APC-Cy7-conjugated mouse anti-human CD14 (BD Bioscience, San Jose, CA), PE-conjugated anti-human CD45, APC-conjugated anti-human CD38 (eBioscience, San Diego, CA). The stained cells were analyzed with on a BD FACS Canto II flow cytometer.
AML xenogeneic mouse model
For the THP1 xenograft model, two-day old NSG pups received 1.3 Gy of irradiation prior to intrahepatic transplantation of 1X106 luciferase-transduced human THP1 cells. The recipients were treated with echinomycin or vehicle by intraperitoneal injection. For TP53-mutated AML xenografts, adult NSG mice received 1.3 Gy of irradiation prior to i.v. injection with 2 to 5X106 of AML cells from AML patients. Human CD45-positive (hCD45+) cells were monitored in the blood of recipients by FACS analysis. When the percentage of hCD45+ cells were detectable by flow cytometry the mice were randomized to receive different treatments. Mice that had not reconstituted human AML cells were excluded. The percentage of hCD45+ cells was monitored by FACS analysis to observe the therapeutic effect. For the splenocyte analysis, mice were sacrificed and splenocytes were collected and stained with antibodies specific for hCD45 and mouse leukocyte markers (mCD45) in conjunction with CD34 and CD38 or with HLA-DR, CD33, CD14, CD11b and CD123 for FACS analysis. For the animal studies, at least three mice per group were used to ensure adequate power to detect difference between groups. For all experiments testing animal survival, at least 5 animals were used per group. Animals were randomized into groups. All animal experiments have been performed at least twice. All animal studies were blinded and conducted under the guidelines of the Institutional Animal Care and Use Committee of the University of Maryland and the Children’s National Medical Center.
Bioluminescence imaging
Luciferase activity at each time point was analyzed in mice anesthetized with isoflurane 10 minutes after intraperitoneal injection of d-luciferin potassium salt (Caliper Life Sciences) at 150 mg/kg. Mice were imaged in a Xenogen IVIS Spectrum Imaging System (Caliper Life Sciences). Living Image software was used to analyze the bioluminescent image data. Total bioluminescent signal was obtained as photons/second and regions of interest were used to calculate regional signals.
PK analysis by LC-MS/MS
NSG mice received a single intravenous dose of echinomycin at 100 ug/kg, and blood samples were collected at different time points after dosing. The plasma fraction was immediately separated and stored at −80°C until analysis. To extract echinomycin from plasma, protein was precipitated from the plasma by mixing with acetonitrile 1:4 (v/v plasma: ACN). Echinomycin was monitored by multiple reaction monitoring (MRM) in the positive electrospray ionization mode on an ABI-5500 Qtrap (Sciex, Ontario, Canada) mass spectrometer in tandem with a Shimadzu high performance liquid chromatography (HPLC) system. The Q1 and Q3 transition of echinomycin (m/z 1101.41053.4) and its collision energy were selected and optimized by direct infusion. Chromatographic separation was achieved on an Agilent Poroshell 120, C18 HPLC column at a flow rate of 0.4 mL/min in 7.5 minutes by a gradient elution of water and acetonitrile containing 0.1% formic acid.
Statistical analyses
Two tailed T tests were used to determine P values for statistical significance of all pair-wise comparisons. For kinetics studies with multiple time points, statistics were analyzed by one-way ANOVA with Tukey’s multiple comparisons test. Mouse survival was estimated by Kaplan-Meier survival analysis, with statistical significance determined by the log-rank test.
Supplementary Material
Acknowledgments
This study was supported by grants from the National Institutes of Health National Cancer Institute [CA171972, CA183030 (Y Liu) and CA164469 (Y Wang)] and a grant from OncoImmune, Inc. Some of the studies were performed when the authors were at the Children’s National Medical Center in Washington DC.
Footnotes
Conflict of Interest Disclosures: Authors disclosed no potential conflicts of interest.
References:
- 1.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2010. January; 2(1): a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isobe M, Emanuel BS, Givol D, Oren M, Croce CM. Localization of gene for human p53 tumour antigen to band 17p13. Nature 1986. March 6–12; 320(6057): 84–85. [DOI] [PubMed] [Google Scholar]
- 3.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science 1991. July 5; 253(5015): 49–53. [DOI] [PubMed] [Google Scholar]
- 4.Haferlach C, Dicker F, Herholz H, Schnittger S, Kern W, Haferlach T. Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia 2008. August; 22(8): 1539–1541. [DOI] [PubMed] [Google Scholar]
- 5.Rucker FG, Schlenk RF, Bullinger L, Kayser S, Teleanu V, Kett H, et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood 2012. March 1; 119(9): 2114–2121. [DOI] [PubMed] [Google Scholar]
- 6.Hou HA, Chou WC, Kuo YY, Liu CY, Lin LI, Tseng MH, et al. TP53 mutations in de novo acute myeloid leukemia patients: longitudinal follow-ups show the mutation is stable during disease evolution. Blood Cancer J 2015; 5: e331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ok CY, Patel KP, Garcia-Manero G, Routbort MJ, Peng J, Tang G, et al. TP53 mutation characteristics in therapy-related myelodysplastic syndromes and acute myeloid leukemia is similar to de novo diseases. J Hematol Oncol 2015; 8: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Yehuda D, Krichevsky S, Caspi O, Rund D, Polliack A, Abeliovich D, et al. Microsatellite instability and p53 mutations in therapy-related leukemia suggest mutator phenotype. Blood 1996. December 1; 88(11): 4296–4303. [PubMed] [Google Scholar]
- 9.Shih AH, Chung SS, Dolezal EK, Zhang SJ, Abdel-Wahab OI, Park CY, et al. Mutational analysis of therapy-related myelodysplastic syndromes and acute myelogenous leukemia. Haematologica 2013. June; 98(6): 908–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wattel E, Preudhomme C, Hecquet B, Vanrumbeke M, Quesnel B, Dervite I, et al. p53 mutations are associated with resistance to chemotherapy and short survival in hematologic malignancies. Blood 1994. November 1; 84(9): 3148–3157. [PubMed] [Google Scholar]
- 11.Bally C, Ades L, Renneville A, Sebert M, Eclache V, Preudhomme C, et al. Prognostic value of TP53 gene mutations in myelodysplastic syndromes and acute myeloid leukemia treated with azacitidine. Leuk Res 2014. July; 38(7): 751–755. [DOI] [PubMed] [Google Scholar]
- 12.Bejar R, Stevenson KE, Caughey B, Lindsley RC, Mar BG, Stojanov P, et al. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2014. September 1; 32(25): 2691–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welch JS, Petti AA, Miller CA, Fronick CC, O’Laughlin M, Fulton RS, et al. TP53 and Decitabine in Acute Myeloid Leukemia and Myelodysplastic Syndromes. The New England journal of medicine 2016. November 24; 375(21): 2023–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blagosklonny MV, An WG, Romanova LY, Trepel J, Fojo T, Neckers L. p53 inhibits hypoxia-inducible factor-stimulated transcription. The Journal of biological chemistry 1998. May 15; 273(20): 11995–11998. [DOI] [PubMed] [Google Scholar]
- 15.Huang J, Zhang Y, Bersenev A, O’Brien WT, Tong W, Emerson SG, et al. Pivotal role for glycogen synthase kinase-3 in hematopoietic stem cell homeostasis in mice. J Clin Invest 2009. December; 119(12): 3519–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birner P, Schindl M, Obermair A, Breitenecker G, Oberhuber G. Expression of hypoxia-inducible factor 1alpha in epithelial ovarian tumors: its impact on prognosis and on response to chemotherapy. Clinical cancer research : an official journal of the American Association for Cancer Research 2001. June; 7(6): 1661–1668. [PubMed] [Google Scholar]
- 17.Wang Y, Liu Y, Malek SN, Zheng P. Targeting HIF1alpha Eliminates Cancer Stem Cells in Hematological Malignancies. Cell Stem Cell 2011. April 8; 8(4): 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Li H, Xi HS, Li S. HIF1alpha is required for survival maintenance of chronic myeloid leukemia stem cells. Blood 2012. March 15; 119(11): 2595–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheloni G, Tanturli M, Tusa I, Ho DeSouza N, Shan Y, Gozzini A, et al. Targeting chronic myeloid leukemia stem cells with the hypoxia-inducible factor inhibitor acriflavine. Blood 2017. August 3; 130(5): 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Liu Y, Malek SN, Zheng P, Liu Y. Targeting HIF1alpha eliminates cancer stem cells in hematological malignancies. Cell stem cell 2011. April 8; 8(4): 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Liu Y, Tang F, Bernot KM, Schore R, Marcucci G, et al. Echinomycin protects mice against relapsed acute myeloid leukemia without adverse effect on hematopoietic stem cells. Blood 2014. August 14; 124(7): 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkin B, Erba H, Ouillette P, Roulston D, Purkayastha A, Karp J, et al. Acquired genomic copy number aberrations and survival in adult acute myelogenous leukemia. Blood 2010. December 2; 116(23): 4958–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018. October; 562(7728): 526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugimoto K, Toyoshima H, Sakai R, Miyagawa K, Hagiwara K, Ishikawa F, et al. Frequent mutations in the p53 gene in human myeloid leukemia cell lines. Blood 1992. May 1; 79(9): 2378–2383. [PubMed] [Google Scholar]
- 25.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 2006. December; 6(12): 909–923. [DOI] [PubMed] [Google Scholar]
- 26.Selivanova G, Iotsova V, Okan I, Fritsche M, Strom M, Groner B, et al. Restoration of the growth suppression function of mutant p53 by a synthetic peptide derived from the p53 C-terminal domain. Nat Med 1997. June; 3(6): 632–638. [DOI] [PubMed] [Google Scholar]
- 27.Kong D, Park EJ, Stephen AG, Calvani M, Cardellina JH, Monks A, et al. Echinomycin, a small-molecule inhibitor of hypoxia-inducible factor-1 DNA-binding activity. Cancer Research 2005. October 1; 65(19): 9047–9055. [DOI] [PubMed] [Google Scholar]
- 28.Corbaz VR, Ettlinger L, Gaumann E, Keller-Schierlein W, Kradolfer F, Neipp L, et al. Metabolic products of actinomycetes: echinomycin. Helv Chim Acta 1957; 40: 199–204. [Google Scholar]
- 29.Iyer AK, Khaled G, Fang J, Maeda H. Exploiting the enhanced permeability and retention effect for tumor targeting. Drug Discov Today 2006. September; 11(17–18): 812–818. [DOI] [PubMed] [Google Scholar]
- 30.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. Journal of controlled release : official journal of the Controlled Release Society 2000. March 1; 65(1–2): 271–284. [DOI] [PubMed] [Google Scholar]
- 31.Cerqueira BB, Lasham A, Shelling AN, Al-Kassas R. Nanoparticle therapeutics: Technologies and methods for overcoming cancer. European journal of pharmaceutics and biopharmaceutics : official journal of Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik eV 2015. November; 97(Pt A): 140–151. [DOI] [PubMed] [Google Scholar]
- 32.Kumar P, Gulbake A, Jain SK. Liposomes a vesicular nanocarrier: potential advancements in cancer chemotherapy. Critical reviews in therapeutic drug carrier systems 2012; 29(5): 355–419. [DOI] [PubMed] [Google Scholar]
- 33.Silverman JA, Deitcher SR. Marqibo(R) (vincristine sulfate liposome injection) improves the pharmacokinetics and pharmacodynamics of vincristine. Cancer chemotherapy and pharmacology 2013. March; 71(3): 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.