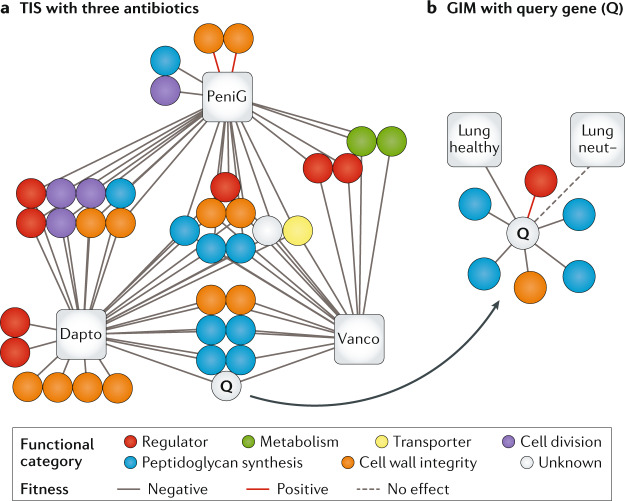
Fig. 4. Mapping complex genotype–phenotype relationships.
a | A gene–antibiotic network for three antibiotics (based on refs54,119). Each node (circle) depicts a gene, whereas each edge indicates a negative (solid grey line), neutral (dashed grey line) or positive (solid red line) effect on fitness between the genes in the presence of antibiotics as determined by transposon-insertion sequencing (TIS). All three antibiotics — penicillin G (PeniG), vancomycin (Vanco) and daptomycin (Dapto) — affect cell wall integrity but TIS uncovers a wide variety of genes involved, many of which are not direct targets of the antibiotic. In this case, the unknown gene Q is likely to be involved in peptidoglycan synthesis/membrane integrity on the basis of the function of other genes with similar fitness profiles. b | By mutation of gene Q and construction of a transposon library in this mutant background, genetic interactions can be identified (based on concepts from ref.33). In this case, the genes uncovered in the mutant background, as depicted in the gene interaction map (GIM), further support a role in peptidoglycan synthesis, that it may function in the cell membrane and that it may be controlled by a particular regulator. Additionally, in vivo TIS data with the mutant library performed in healthy mice and mice depleted of neutrophils (neut−) indicate that gene Q is needed to establish lung infection but is dispensable in the absence of neutrophils. Adapted from ref.54, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).