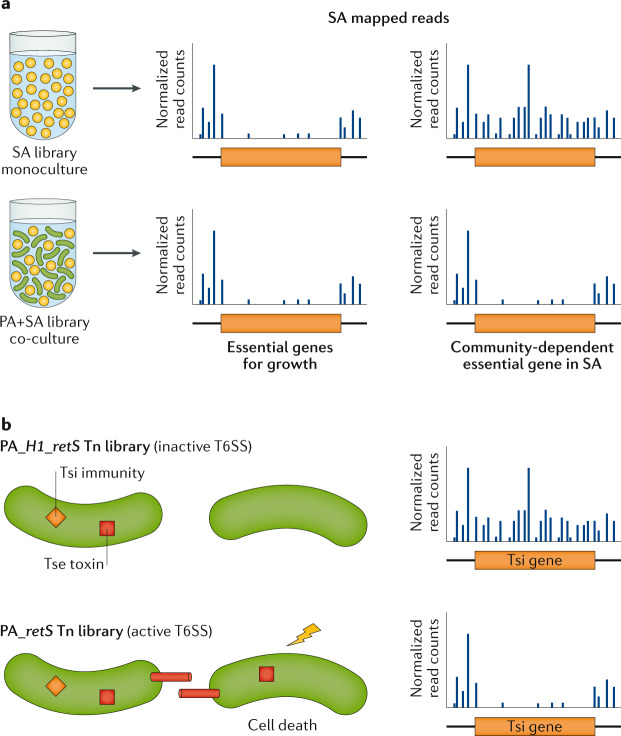
Fig. 6. TIS to assay microorganism–microorganism interactions.
a | Transposon-insertion sequencing (TIS) of Staphylococcus aureus (SA) mutants alone (orange circles) compared with coculture of SA and Pseudomonas aeruginosa (PA) wild-type strain (green rods) can identify community-dependent essential genes that are needed only during coculture. Other features are as in Fig. 1. b | Using TIS to identify type VI secretion system (T6SS) toxin immunity pairs by growing cells with close cell-to-cell contact, and performing TIS so that genes involved in protection of T6SS-depending killing (depicted as a yellow bolt) can be detected. In this example, genes in PA encoding immunity proteins (Tsi; orange diamond) that protect from neighbour killing by toxins (Tse; red square) become essential only in a T6SS-active (retS; bottom panel) background but not in the inactive (H1_retS; top panel) cellular background.